IIK Research Group Microbial Genome Analysis
- Whole Genome Sequencing of Hospital-Relevant Pathogens -About us
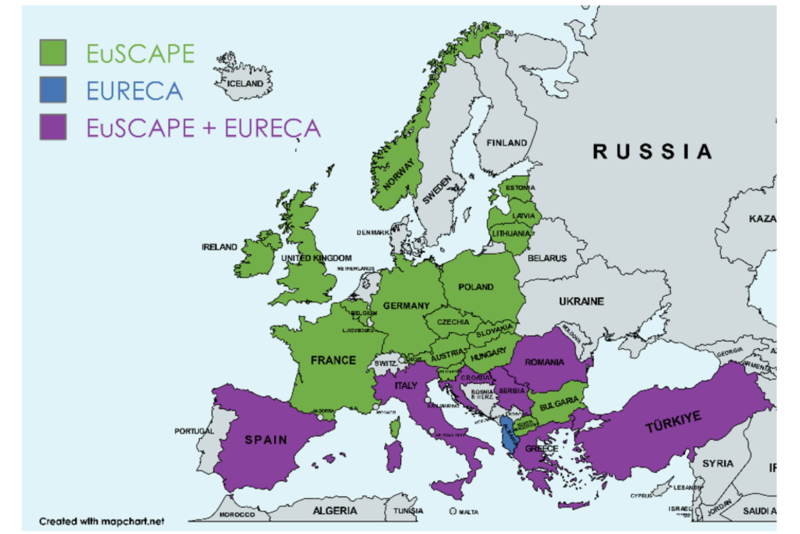
We use whole genome sequencing (WGS) to look at the very makings of each bacterium. By deducing its genetic code we have the ultimate resolution to inform us about putative transmission events on hospital wards, which allows us to investigate outbreaks. Furthermore, we can use the data generated to learn about antibiotic resistance. We are also involved in a number of projects with internal and external collaborators. One of our main interests is the spread of Gram-negative multidrug resistant clones through hospital networks. For this, we collaborate closely with colleagues at the Wellcome Trust Sanger Institute (Cambridge, UK), Big Data Institute (Oxford, UK), University Medical Center Groningen (Groningen, Netherlands), Charité Berlin (Berlin, Germany), and Justus-Liebig-University Hospital (Gießen, Germany).
Selected Publications
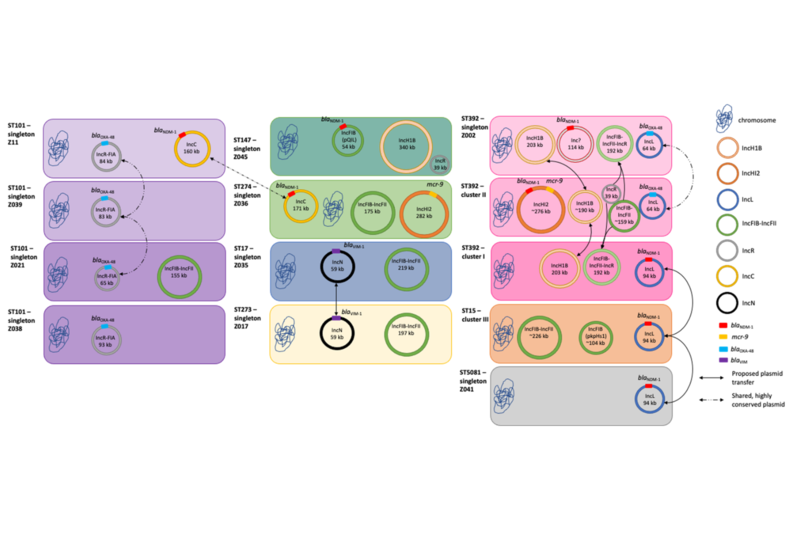
- Akintayo I, et al. Tracking clonal and plasmid transmission in colistin- and carbapenem-resistant Klebsiella pneumoniae. mSystems 2025 10(2):e01128-24.
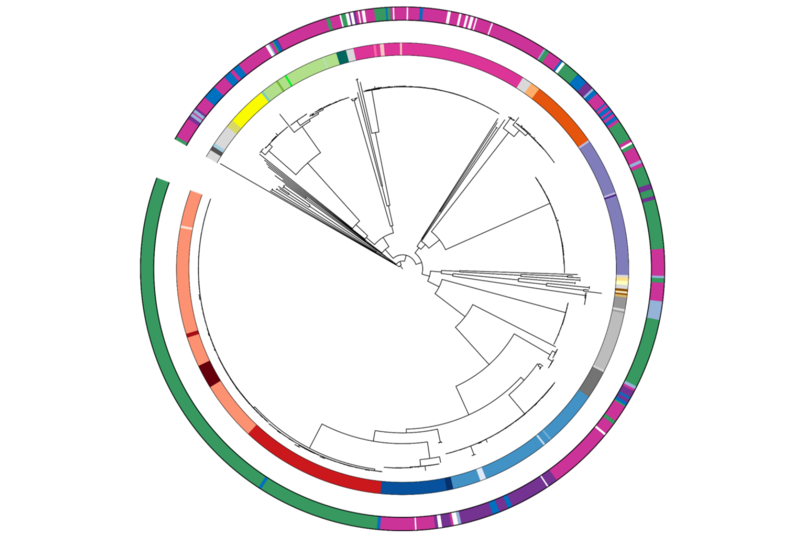
- Budia-Silva M, et al. International and regional spread of carbapenem-resistant Klebsiella pneumoniae in Europe. Nature Communications 2024 15(1):5092.
- Reuter S, et al. A global view on carbapenem-resistant Acinetobacter baumannii. mBio 2023 14(6):e02260-23.
Technology
- Illumina MiSeq & Oxford Nanopore Technology GridION
- Expertise in bacterial whole genome sequencing and analysis
- Bioinformatics expertise
Funded by
Dr. biol. Sandra Reuter
Tel.: +49 (0) 761 270 82350
Fax: +49 (0) 761 270 83030