Single Cell Immunobiology Group
Dr. Sagar (Group leader)
+49 (0) 761 270 36330
University Medical Center Freiburg
Department of Medicine II
Gastroenterology, Hepatology,
Endocrinology, and Infectious Diseases
Hugstetter Straße 55, 79106 Freiburg
Research Topics
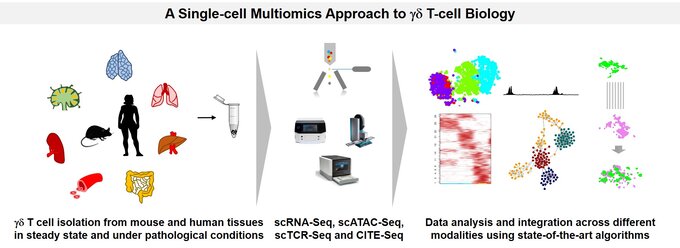
Understanding the development and function of tissue-resident γδ T cells in mice and humans using an integrated single-cell multimodal approach
In mammals studied so far, the majority of T cells express αβ T cell receptor (TCR) on their cell surface and hence these cells are called αβ T cells, for e.g., CD8+ cytotoxic and CD4+ helper T cells. A small subset of T cells expresses a distinct γδ TCR on their cell surface and are called γδ T cells. Although a significant amount of research has focused on understanding the development and the functions of αβ T cells in homeostasis and disease, the γδ lineage is relatively understudied. The γδ T cell lineage has sparked a significant recent interest among immunologists because of their critical roles in infection, tumor growth and autoimmune diseases and is being examined in various cancer immunotherapy trials.
In our group, we study the role of γδ T cells in tissue physiology and pathology (such as inflammatory bowel disease) at a genome-wide single-cell resolution. We employ cutting-edge single-cell experimental methods such as single-cell RNA sequencing, CITE-Seq, single-cell TCR repertoire sequencing and single-cell ATAC-Seq to profile γδ T cells across different tissues in humans and mice in steady state as well as immunopathology to underpin the molecular mechanisms governing their modes of action. A special focus of the lab is also to perform extensive integrated comparative analysis across different modalities, tissues and species using state-of-the-art single-cell computational methodologies to identify cell states and the underlying transcriptional and epigenetic programs which govern their identities at single-cell resolution.
Combining the power of highly-resolved single-cell technologies with mouse models and human samples from various immune diseases, we aim to dig deeper and acquire an improved understanding of the underlying mechanisms of γδ T cell functions and the nature of their interaction with other immune and non-immune cell types in homeostasis and pathologies such as inflammatory bowel disease and viral hepatitis in close collaboration with the group of Maike Hofmann and Robert Thimme.
Methodology
- Preparation of single-cell suspension from various murine and human tissues
- Fluorescence-activated cell sorting (FACS)
- Single-cell profiling (mRNA, chromatin accessibility and cell surface markers) using 384 well plate-based (using liquid handlers) and 10x Genomics platform
- in vitro cellular differentiation and activation assays
- Single molecule fluorescence in situ hybridization (smFISH) and spatial transcriptomics
- Extensive single-cell computational data analysis including methods to integrate datasets across different modalities, tissues and species
- Sagar, Pokrovskii M, Herman JS, Naik S, Sock E, Zeis P, Lausch U, Wegner M, Tanriver Y, Littman DR, Grun D (2020) Deciphering the Regulatory Landscape of gd T Cell Development by Single-Cell RNA-Sequencing. EMBO Je104159
- Sagar, Grun D (2020) Deciphering Cell Fate Decision by Integrated Single-Cell Sequencing Analysis. Annual Review of Biomedical Data Science, 3, 1-22
- Aizarani N, Saviano A*, Sagar*, Mailly L, Durand S, Herman JS, Pessaux P, Baumert TF, Grun D (2019) A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572: 199-204
- Sagar, Herman JS, Pospisilik JA, Grun D (2018) High-Throughput Single-Cell RNA Sequencing and Data Analysis. Methods in Molecular Biology 1766: 257-283
- Herman JS, Sagar, Grun D (2018) FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nature Methods 15: 379-386

We are part of the TRANSLATIONAL EXPERIMENTAL IMMUNOLOGY LAB
Prof. Dr. Maike Hofmann
Head of the Translational Experimental Immunology Section
Head of Research MED II
+49 (0) 761 270-35091
Prof. Dr. Dr. Bertram Bengsch
Head of the Translational Systems Immunology Section in Hepatogastroenterology
+49 (0) 761 270-32870