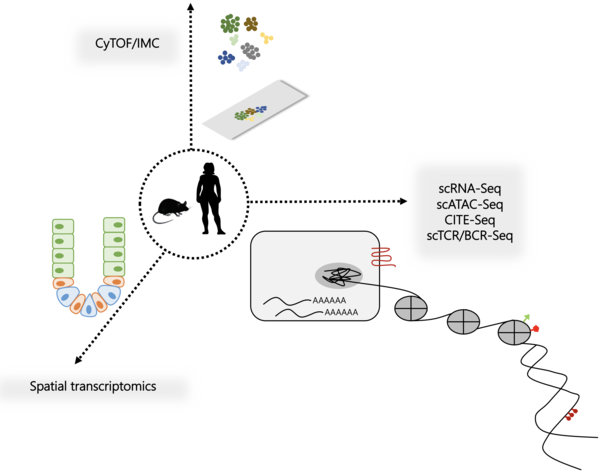
Single-cell Omics Platform
The cell is the basic functional unit of the body and key to understanding health and disease. Recent technological advances now enable in-depth profiling of single cells required for the multi-dimensional understanding of cellular biology in a high-throughput fashion. In the Freiburg single-cell omics unit, we concentrate interdisciplinary expertise across multiple single-cell techniques to drive systems-level understanding at the single cell level. A major goal is to advance the insights into cellular biology for both basic and translational research questions, refining the single-cell methodology and providing optimized pipelines for thorough analysis, thus ultimately accelerating single-cell multiomics projects in a time and resource efficient way.
| Vision | Technologies |
|---|---|
|
|
Prof. Dr. Dr. Bertram Bengsch
Dr. Gianni Monaco
Dr. Maike Hofmann
Dr. Roman Sankowski
Dr. Sagar
Klinik für Innere Medizin II
Hugstetter Straße 55, 79106 Freiburg
Institut für Neuropathologie
Breisacher Straße 64, 79106 Freiburg
Hensel N, Gu Z, Sagar, Wieland D, Jechow K, Kemming J, Llewellyn-Lacey S, Gostick E, Sogukpinar O, Emmerich F, Price DA, Bengsch B, Boettler T, Neumann-Haefelin C, Eils R, Conrad C, Bartenschlager R, Grün D, Ishaque N, Thimme R, Hofmann M. Memory-like HCV-specific CD8+ T cells retain a molecular scar after cure of chronic HCV infection. Nat Immunol. 2021 Feb;22(2):229-239. doi: 10.1038/s41590-020-00817-w.
Schulien I, Kemming J, Oberhardt V, Wild K, Seidel LM, Killmer S, Sagar, Daul F, Salvat Lago M, Decker A, Luxenburger H, Binder B, Bettinger D, Sogukpinar O, Rieg S, Panning M, Huzly D, Schwemmle M, Kochs G, Waller CF, Nieters A, Duerschmied D, Emmerich F, Mei HE, Schulz AR, Llewellyn-Lacey S, Price DA, Boettler T, Bengsch B, Thimme R, Hofmann M, Neumann-Haefelin C. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat Med. 2021 Jan;27(1):78-85. doi: 10.1038/s41591-020-01143-2.
Sagar, Pokrovskii M, Herman JS, Naik S, Sock E, Zeis P, Lausch U, Wegner M, Tanriver Y, Littman DR, Grün D. Deciphering the regulatory landscape of fetal and adult γδ T-cell development at single-cell resolution. EMBO J. 2020 Jul 1;39(13):e104159. doi: 10.15252/embj.2019104159.
Masuda T, Amann L, Sankowski R, Staszewski O, Lenz M, D Errico P, Snaidero N, Costa Jordão MJ, Böttcher C, Kierdorf K, Jung S, Priller J, Misgeld T, Vlachos A, Meyer-Luehmann M, Knobeloch KP, Prinz M. Novel Hexb-based tools for studying microglia in the CNS. Nat Immunol. 2020 Jul;21(7):802-815. doi: 10.1038/s41590-020-0707-4.
Sagar, Grün D. Deciphering Cell Fate Decision by Integrated Single-Cell Sequencing Analysis. Annu Rev Biomed Data Sci. 2020 Jul;3:1-22. doi: 10.1146/annurev-biodatasci-111419-091750.
Masuda T, Sankowski R, Staszewski O, Prinz M. Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 2020 Feb 4;30(5):1271-1281. doi: 10.1016/j.celrep.2020.01.010.
Mereu E, Lafzi A, Moutinho C, Ziegenhain C, McCarthy DJ, Álvarez-Varela A, Batlle E, Sagar, Grün D, Lau JK, Boutet SC, Sanada C, Ooi A, Jones RC, Kaihara K, Brampton C, Talaga Y, Sasagawa Y, Tanaka K, Hayashi T, Braeuning C, Fischer C, Sauer S, Trefzer T, Conrad C, Adiconis X, Nguyen LT, Regev A, Levin JZ, Parekh S, Janjic A, Wange LE, Bagnoli JW, Enard W, Gut M, Sandberg R, Nikaido I, Gut I, Stegle O, Heyn H. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol. 2020 Jun;38(6):747-755. doi: 10.1038/s41587-020-0469-4.
Sagar, Grün D. Lineage Inference and Stem Cell Identity Prediction Using Single-Cell RNA-Sequencing Data. Methods Mol Biol. 2019;1975:277-301. doi: 10.1007/978-1-4939-9224-9_13.
Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar, Sindram E, Seredenina T, Muhs A, Scheiwe C, Shah MJ, Heiland DH, Schnell O, Grün D, Priller J, Prinz M. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat Neurosci. 2019 Dec;22(12):2098-2110. doi: 10.1038/s41593-019-0532-y.
Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Grün D, Priller J, Stadelmann C, Prinz M. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019 Feb;566(7744):388-392. doi: 10.1038/s41586-019-0924-x.
Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, Pessaux P, Baumert TF, Grün D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019 Aug;572(7768):199-204. doi: 10.1038/s41586-019-1373-2.
Jordão MJC, Sankowski R, Brendecke SM, Sagar, Locatelli G, Tai YH, Tay TL, Schramm E, Armbruster S, Hagemeyer N, Groß O, Mai D, Çiçek Ö, Falk T, Kerschensteiner M, Grün D, Prinz M. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019 Jan 25;363(6425):eaat7554. doi: 10.1126/science.aat7554.
Bengsch B, Ohtani T, Khan O, Setty M, Manne S, O'Brien S, Gherardini PF, Herati RS, Huang AC, Chang KM, Newell EW, Bovenschen N, Pe'er D, Albelda SM, Wherry EJ. Epigenomic-Guided Mass Cytometry Profiling Reveals Disease-Specific Features of Exhausted CD8 T Cells. Immunity. 2018 May 15;48(5):1029-1045.e5. doi: 10.1016/j.immuni.2018.04.026.
Sagar, Herman JS, Pospisilik JA, Grün D. High-Throughput Single-Cell RNA Sequencing and Data Analysis. Methods Mol Biol. 2018;1766:257-283. doi: 10.1007/978-1-4939-7768-0_15.
Tay TL, Sagar, Dautzenberg J, Grün D, Prinz M. Unique microglia recovery population revealed by single-cell RNAseq following neurodegeneration. Acta Neuropathol Commun. 2018 Sep 5;6(1):87. doi: 10.1186/s40478-018-0584-3.
Herman JS, Sagar, Grün D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat Methods. 2018 May;15(5):379-386. doi: 10.1038/nmeth.4662.