Molecular biology of bornaviruses
Bornaviruses exhibits several interesting biological features that render these viruses unique members of the order Monanegavirales. Borna disease virus (BDV), the prototype family member, has adopted an elaborate mechanism for removing nucleotides from the 5’end of viral genomic RNAs and re-acquiring the missing genetic information by elongating the genomes on internal template motifs after realignment of the 3’ termini (see figure). Since bornaviruses are very poor inducers of interferon, we hypothesize that bornaviruses have evolved this complex replication strategy to efficiently remove 5’-triphosphates from the genome which otherwise would be sensed by pattern recognition receptors of infected cells.

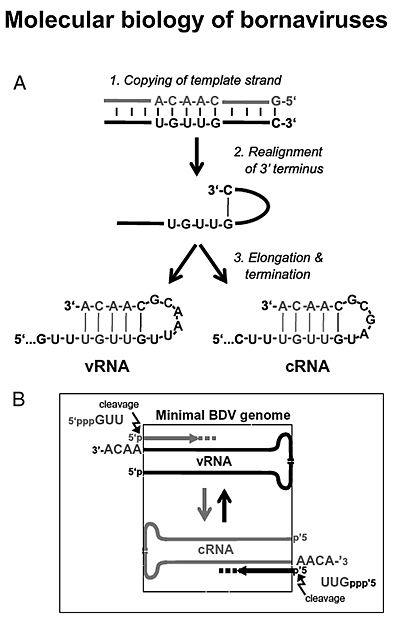
Model of the terminal realign-and-elongation process and its presumed function in BDV infection.
(A) Schematic representation of the realign-and-elongation process responsible for the terminal elongation of BDV v- and cRNA. (B) Schematic representation of the proposed function of terminal elongation in BDV replication. The synthesis of 3′nucleotides from internal templates permits synthesis and enzymatic removal of 5′ nucleotides carrying the triphosphate groups without loss of genetic information. Virus RNA and cRNA molecules are shown in black and gray colors, respectively. The dashed box frames the minimal genetic information required for the establishment of BDV replication. (Figure from Martin et al. 2011)
Open questions presently being investigated include:
a) Nature of endonuclease responsible for trimming of 5’ ends of BDV genomic RNAs.
b) Conservation of viral genome trimming mechanism in related viruses, including avian bornaviruses and Nyamanini virus.
Selected Publications from our group
- Schneider U, Schwemmle M, Staeheli P. Genome trimming: a unique strategy for replication control employed by Borna disease virus. Proc. Natl. Acad. Sci. USA 9: 3441-3446 (2005)
- Schneider U, Martin A, Schwemmle M, Staeheli P. Genome trimming by Borna disease viruses: Viral replication control or escape from cellular surveillance? Cell Mol Life Sci. 64: 1038-1042 (2007)
- Habjan M, Andersson I, Klingström J, Schümann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Mühlberger E, Mirazimi A, Weber F. Processing of genomic 5’ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3: e2032 (2008)
- Reuter A, Ackermann A, Kothlow S, Rinder M, Kaspers B, Staeheli P. Avian bornaviruses escape recognition by the innate immune system. Viruses 2: 927-938 (2010)
- Martin A, Hoefs N, Tadewaldt J, Staeheli P, Schneider U. Modification of viral genomic RNA by terminal elongation on internal template motifs. Proc. Natl. Acad. Sci. USA 108: 7206-7211 (2011)
