Projects
In the ERAPerMed project PARADISE, focusing on ANCA vasculitis as an exemplary relapsing multi-system autoimmune disease, we use patient level data to identify the characteristics, or markers, most often linked to relapse events.
See more: https://paradise-project.eu
Responsible: Moritz Hess
Collaborators: Harald Binder, Maren Hackenberg, Paradise Consortium
This project currently focuses on the application of deep neural networks, mainly generative models, and mathematical models to explore innovative solutions in medicine. I aim to combine these approaches to enhance our understanding of complex medical phenomena and support advancements in healthcare.
See more: https://www.smalldata-initiative.de/projects/c03/
Responsible: Hanning Yang, Moritz Hess
Collaborators: Cristina Has, Meropi Karakioulaki
Parameter optimization in ODE models remains challenging due to limited experimental data and the complexity of biological systems. This project aims to improve performance and reliability by benchmarking existing methods and using reinforcement learning to identify and adapt optimal optimization strategies.
See more: https://www.smalldata-initiative.de/projects/c03/
Responsible: Niklas Neubrand, Moritz Hess
Collaborators: Jens Timmer, Yaser Kord
We aim to recycle sawdust back into solid wood blocks by organically crosslinking proteins in the wood with bacteria genetically designed for this task. Our group specifically aims to integrate classical experimental design techniques with modern AI prediction algorithms such as TabPFN to streamline materials design and navigate all the possible combinations of material composition and production processes.
Responsible: Tim Litwin
Collaborators: Wilfried Weber, Thomas Speck
In the context of our statistical consultation of animal experiments, we compare the original statistical design and analysis plan with the actually conducted analyses published in the final study to gather insights about which statistical advices get lost during the conduct of the study.
Responsible: Claire Reiland, Tim Litwin
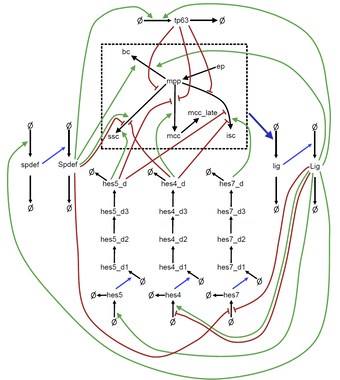
This project aims at understanding the cellular mechanisms that dictate cell fate decisions in the formation of mucociliary cells in Xenopus embryos. We contribute to the functional understanding of this developmental process by formulating a mathematical model based on ordinary differential equations, which provide a mechanistic explanation of how Notch signaling induces the correct cell type specifications.
Responsible: Tim Litwin
Collaborators: Peter Walentek
Data2Dynamics (D2D) is an open source MATLAB toolbox for mathematical modeling primarily used in systems biology and is tailored to parameter estimation and uncertainty analysis in dynamical systems.
D2D is further developed and new features are intergrated. Recent Additions from our group:
- The dose-dependent Retarded Transient Function (RTF) as an alternative to ODE modelling and a tool for dimensionality reduction in ODE systems.
- Interface for the supporting software for automated model selection PEtab Select.
Responsible: Tim Litwin, Niklas Neubrand, Timo Rachel
Collaborators: Jens Timmer, Andreas Raue, Clemens Kreutz