Structure-Based Drug Discovery
targeting novel oncogenic methyltransferases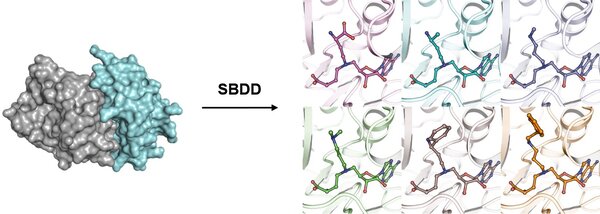
Epigenetic dysregulation, particularly aberrant methylation, profoundly influences cancer development and progression. The methyltransferase (MTase) family, which catalyze the transfer of methyl groups to DNA, RNA, and protein substrates, has emerged as a master regulator in cancer pathogenesis. These enzymes exert critical control over diverse biological functions, including DNA repair, cell cycle progression, transcriptional regulation, RNA processing, protein translation and stability. Given the widespread dysregulation of MTase activity in cancer, this enzyme class represents a highly attractive and clinically validated set of therapeutic targets. Indeed, the successful development and clinical advancement of small molecule inhibitors targeting enzymes such as DNMTs, EZH2, PRMT5, and METTL3 underscore the profound therapeutic potential inherent in modulating this family of enzymes.
Despite these advances, a substantial knowledge gap persists. Among the more than 200 putative MTase encoded in the human genome, a significant proportion remains functionally uncharacterized and critically lacks the requisite chemical tools for rigorous interrogation. This absence of potent and selective chemical probes constitutes a major bottleneck to advancing the field and, simultaneously, presents a profound opportunity for therapeutic innovation. Such probes are indispensable as a foundational step to rigorously validate novel targets, elucidate their biological functions, and thereby forge a direct pathway toward the development of next-generation cancer therapeutics.
Our lab has established and successfully implemented an integrated pipeline for the discovery and rigorous validation of chemical probes against novel and understudied MTase targets. Our track record demonstrates the efficacy of this approach, exemplified by our development of the first-in-class inhibitor and degrader for KMT9, the first-in-class inhibitor targeting a novel histone methyltransferase KMT10 (Manuscript in preparation), and an advanced project focused on another unexploited protein lysine methyltransferase. This pipeline integrates interdisciplinary methodologies, spanning from multi-omics data mining for target prioritization to structural biology, rational structure-based drug design, and a stringent preclinical validation cascade.
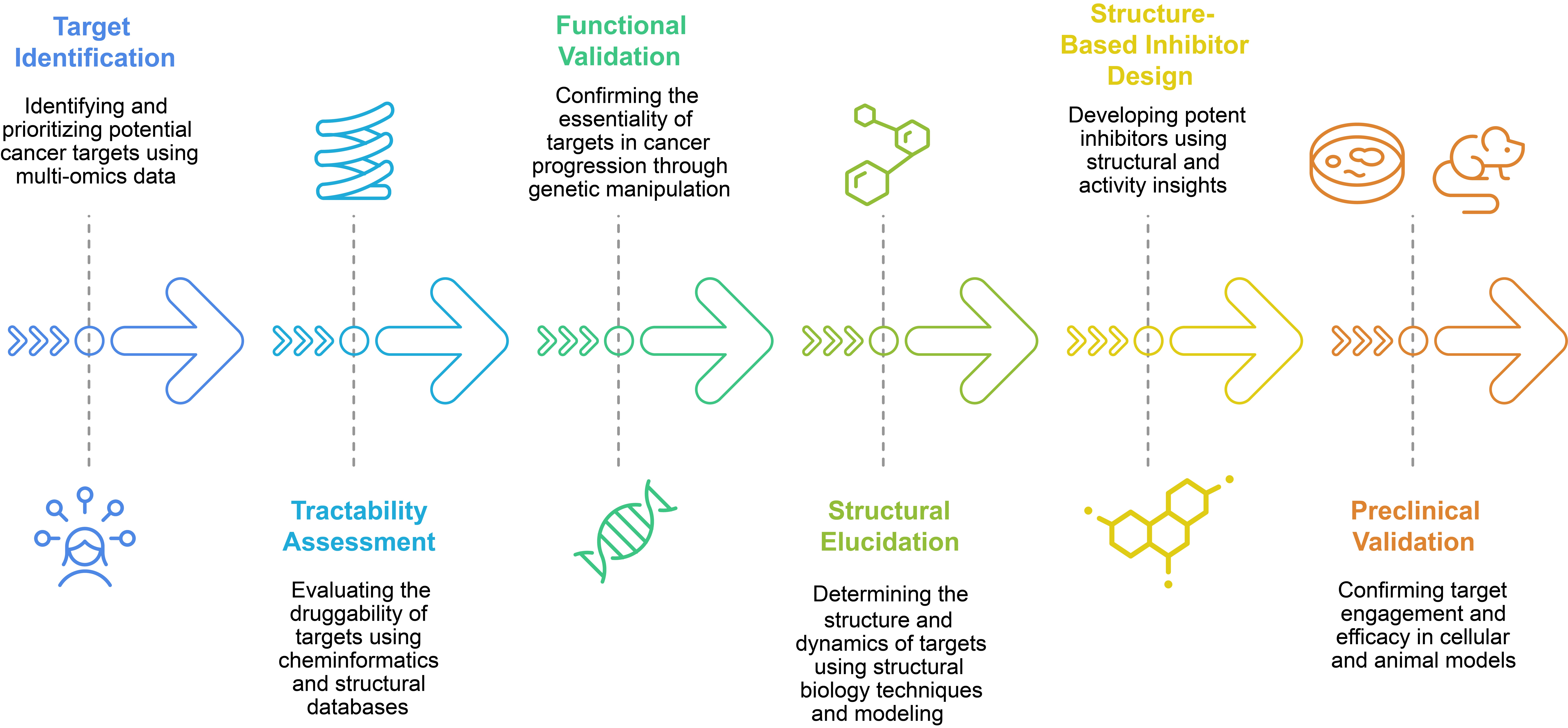
Target Identification & Prioritization: We integrate knowledge-based data mining (literature, expertise) with multi-omics analysis (TCGA, DepMap, etc.) to identify understudied methyltransferases with oncogenic potential. This is followed by a rigorous tractability assessment using cheminformatics, structural databases, and AI modeling to prioritize targets for chemical probe development.
Functional & Structural Elucidation: For prioritized targets, we establish foundational knowledge through a two-pronged approach. First, we perform functional validation via genetic perturbation (KD/KO/OE) in cancer cell lines to confirm target essentiality in proliferation, migration, and invasion. Second, we employ state-of-the-art structural biology (X-ray, Cryo-EM, AI modeling, MD simulations) to gain atomic-level mechanistic insights and delineate druggable pockets.
Structure-Based Chemical Probe Development: Leveraging these insights, we execute rational structure-based drug design campaigns, including virtual screening and iterative chemical optimization, to develop potent and selective small-molecule inhibitors or degraders. This involves rapid design-make-test-analyze cycles, with rigorous characterization of hit compounds for potency, selectivity, and mechanism of action using biophysical and enzymatic assays.
Rigorous Preclinical Validation: Promising chemical probes undergo a comprehensive preclinical validation cascade. This includes confirming cellular target engagement (CETSA), demonstrating modulation of downstream pathways and anti-proliferative/migratory/invasive activity (MTT/CellTiter-Glo), and profiling global transcriptional effects (RNA-seq). Finally, lead compounds with optimal drug-like properties are advanced into in vivo efficacy studies using PDX or genetically engineered mouse models for ultimate proof-of-concept.

Dr. Sheng Wang
Center for Clinical Research
University Freiburg Medical Center
Breisacherstrasse 66
D-79106 Freiburg i. Br.
Germany
+49 (0)761-270 63340

Dr. Sylvia Urban
Center for Clinical Research
University Freiburg Medical Center
Breisacherstrasse 66
D-79106 Freiburg i. Br.
Germany
+49 (0)761-270 63360

Dr. Ling Peng
Center for Clinical Research
University Freiburg Medical Center
Breisacherstrasse 66
D-79106 Freiburg i. Br.
Germany
+49 (0)761-270 63910
Literature
Metzger, E.*; Wang, S.*; Urban, S.; Willmann, D.; Schmidt, A.; Offermann, A.; Allen, A.; Sum, M.; Obier, N.; Cottard, F.; Ulferts, S.; Preca, B. T.; Hermann, B.; Maurer, J.; Greschik, H.; Hornung, V.; Einsle, O.; Perner, S.; Imhof, A.; Jung, M.; Schüle, R. KMT9 Monomethylates Histone H4 Lysine 12 and Controls Proliferation of Prostate Cancer Cells. Nat. Struct. Mol. Biol. 2019, 26 (5), 361–371.
Wang, S.; Klein, S. O.; Urban, S.; Staudt, M.; Barthes, N. P. F.; Willmann, D.; Bacher, J.; Sum, M.; Bauer, H.; Peng, L.; Rennar, G. A.; Gratzke, C.; Schüle, K. M.; Zhang, L.; Einsle, O.; Greschik, H.; MacLeod, C.; Thomson, C. G.; Jung, M.; Metzger, E.; Schüle, R. Structure-Guided Design of a Selective Inhibitor of the Methyltransferase KMT9 with Cellular Activity. Nat. Commun. 2024, 15 (1), 43.
Wang, S.; Barthes, N. P. F.; Urban, S.; Hazai, V. I.; Klein, S. O.; Pappert, T.; Kümmel, P.; Heller, N.; Bacher, J.; Staudt, M.; Ruprecht, J.; Peng, L.; Sum, M.; Berlin, C.; Sarraf, D.; Regenass, P.; Warstat, R.; Walz, J.; Mishra, P.; Zhang, L.; Einsle, O.; Günther, S.; Breit, B.; Metzger, E.; Jung, M.; Schüle, R. Structure-Guided Design of a KMT9 Inhibitor Prodrug with Cellular Activity. J. Med. Chem. 2025, 68 (13), 13295–13320.
Schuele, R.; Metzger, E.; Wang, S.; Jung, M.; Klein, S. Specific Small Molecule Inhibitors That Block KMT9 Methyltransferase Activity and Function, WO 2023/017152 A1, 2023.
Schuele, R.; Metzger, E.; Wang, S.; Jung, M.; Barthes, N.; Breit, B.; Sarraf, D.; Pappert, T. Novel Histone Methyltransferase Inhibitors. WO 2021/053158 A1, 2021.
Schuele, R.; Metzger, E.; Wang, S.; Jung, M.; Barthes, N.; Sarraf, D.; Pappert, T. Inhibition of Histone Methyltransferases to Treat Cancer. WO 2020/058358 A1, 2020.
