Research Group „Integrative ImmunoResearch Lab”
Prof. Dr. Michele ProiettiThe AG Proietti is dedicated to understanding why some immune systems struggle to control inflammation or combat infections. By combining experimental and computational approaches, it aims to uncover the underlying mechanisms of immune dysfunction and advance fundamental knowledge in immunology.
Computational work
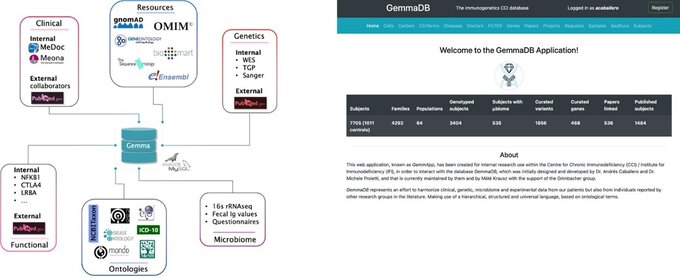
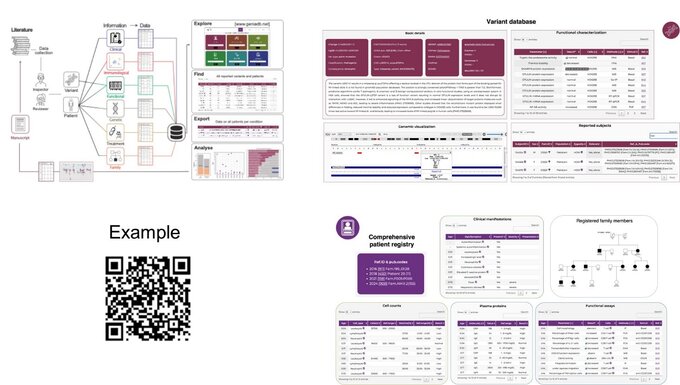
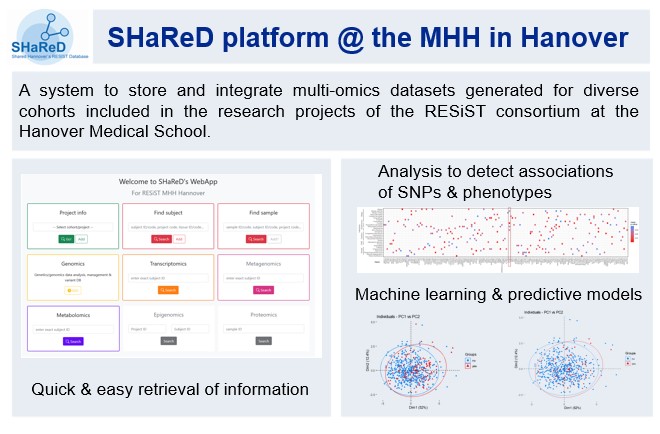
Here, we focus on genetic and genomic data analysis and the development of tools, databases, and applications for research data management and analysis. Our work aims to improve the genetic diagnosis in patients with suspected inborn errors.
We have several expertise that allow us to perform a wide range of bioinformatics tasks, from raw data processing to advanced analysis and visualization, while also developing custom software tools and databases to support their research.
Over the years, we have developed:
Experimental work
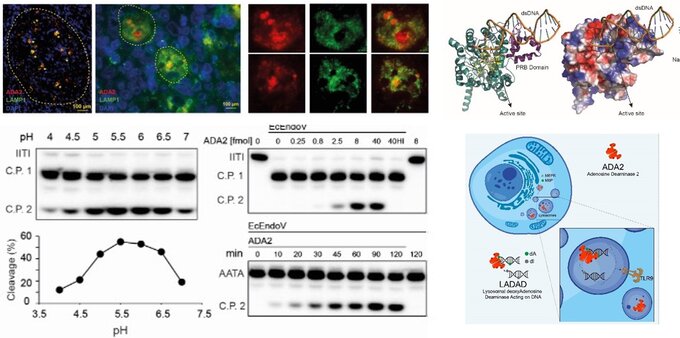
Here, we investigate how how genetic factors and sensing of nucleotides or nucleic acids regulate the human immune response. Recently, we have discovered a new and unexplored pathway related to nucleic acid sensing and immune response. We have found that ADA2 is a lysosomal protein that binds DNA and efficiently deaminates deoxyadenosine (dA) residues of DNA to deoxyinosine (dI), suggesting that DNA is its natural substrate. We also found that dA-to-dI editing of DNA and ADA2 facilitates immune detection of DNA mediated by Toll-like receptor 9 (TLR9). By elucidating ADA2’s role in DNA sensing and metabolism, this study expands our understanding of innate immunity and sheds light on potential therapeutic targets for autoimmune and autoinflammatory diseases. This finding opens up new avenues for understanding immune diseases and potentially developing novel therapeutic approaches (https://www.cell.com/cell-reports/fulltext/S2211-1247(24)01250-6).
Lysosomal deoxyadenosine Deaminase Acting on DNA (LADAD): Our study revealed that ADA2 localizes within the lysosomes, where it interacts with DNA molecules, altering their sequences by converting deoxyadenosine (dA) to deoxyinosine (dI). This activity is crucial in regulating lysosomal immune sensing of nucleic acids (NAs). These results describe a may impact treatment and management of ADA2 deficiency (DADA2) as well as other diseases like autoimmune diseases, cancer, or infectious diseases.
CV
Current position
- Professor (W1), Genetic of Immune Dysregulation, MHH Hannover Medical School
- Head of the Genetic and Genomic Research Unit, Institute for Immunodeficiency (IFI), Medical Center and Faculty of Medicine, University of Freiburg
- Group leader Institute for Immunodeficiency (IFI), Medical Center and Faculty of Medicine, University of Freiburg and Department of Rheumatology and Clinical Immunology, MHH Hannover
Professional career
| Since 2020 | Professor (W1), Genetic of Immunedysregulation, MHH Hannover, Germany |
| Since 2017 | Head of the Genetic and Genomic Research Unit, Institute for Immunodeficiency (IFI), Faculty of Medicine, University of Freiburg. Research focus: the unit acts as a link between clinical and experimental immunologists with the aim of transferring genetic findings into clinic. |
| Since 2017 | Group leader: Institute for Immunodeficiency (IFI), Faculty of Medicine, University of Freiburg. Research focus: genetic and immune-dysregulation. |
| 2015-2017 | Scientist, CCI, Uniklinik Freiburg, Freiburg im Breisgau (Germany). Research focus: genetic and immune-dysregulation. |
| 2010-2015 | Postdoc, Institute of Research in Biomedicine (IRB), Bellinzona, Switzerland. Research focus: purinergic signaling and immune response. |
| 2007-2010 | Clinician, Department of Internal Medicine, University of Genova, Italy. PhD in clinical and experimental Immunology, Centre of Excellence for Biomedical Research (CEBR), University of Genova, Italy. Clinical activity: diagnosis, treatment and care of individuals with autoimmune diseases (e.g. systemic sclerosis, systemic lupus erythematosus, vasculitis, rheumatoid arthritis) and primary immunodeficiencies. Research activity: purinergic signaling and immune response. |
| 2001-2006 | Residency in Internal Medicine, department of internal medicine and clinical immunology unit, University of Rome “La Sapienza”. Clinical training: diagnosis, treatment, and care of patients with complex and multisystem disorders, with particular focus on autoimmune diseases. Research activity: endothelial dysfunction in systemic sclerosis. |
Academic qualifications
- 2010: PhD Clinical and Experimental Immunology, University of Genova, Italy
- 2006: Board Certification in internal Medicine, University of Rome “La Sapienza”
- 2001: Medical degree, University of Rome “La Sapienza”
University training and degree
- 1996-2001: Medical studies, University of Rome “La Sapienza”
- 1999-2001: Clinical Immunology and Allergology Unit, University of Rome “La Sapienza”
Collaborations
National Collaborations:
- Bodo Grimbacher, Center for Chronic Immunodeficiency (CCI)
- Torsten Witte, MHH Hannover, Klinik für Rheumatologie und Immunologie
- Doris Steinemann, MHH Hannover, Institut für Humangenetik
- Nataliya DiDonato, MHH Hannover, Institut für Humangenetik
- Philipp Henneke, UNIVERSITY MEDICAL CENTER and FACULTY OF MEDICINE FREIBURG, Institute for Infection Prevention and Control & Institute for Immunodeficiency
- Stephan Rossart, Uniklinikum Erlangen
- Eva Bartok, UNI Bonn
International Collaborations:
- Ingrun Alseth, Oslo University Hospital
- Ole K Greiner Tollersrud, UiT The Arctic University of Norway.
- Max Warncke, Novartis Institute for Biomedical Research
- Elisabetta Traggiai, Novartis Institute for Biomedical Research
Funding
- CRC/TRR 359 Perinatal Development of Immune Cell Topology (PILOT)
- Cluster of Excellence RESIST (EXC 2155)
- Flex Funding, Cluster of Excellence RESIST
Team
| Group Leader | ||
| Michele Proietti | michele.proietti@uniklinik-freiburg.de | 270-77561 |
| Post-Docs | ||
| Andrés Caballero | andres.caballero@uniklinik-freiburg.de | 270-77729 |
| PhD Students | ||
| Andreas Goschin | andreas.goschin@uniklinik-freiburg.de | |
| MD Students | ||
| David Fuchs | ||
| Natalia Hammersmith |

Center for Chronic Immunodeficiency
at Center for Translational Cell Research
Breisacher Str. 115
79106 Freiburg
Germany