Schilling Laboratory
Welcome to the laboratory of Professor Dr. Oliver Schilling.
The Schilling laboratory pursues a research program in two areas: translational proteomics and proteolytic signaling.

Professor Dr. Oliver Schilling
Heisenberg Professor of Translational Proteomics
Institute of Surgical Pathology
Faculty of Medicine - University of Freiburg
Breisacher Strasse 115a
D-79106 Freiburg
Germany
Tel: +49 761 270 80610
Fax: +49 761 270 81970
oliver.schilling@uniklinik-freiburg.de
oliver.schilling@mol-med.uni-freiburg.de
Students
We are always happy to hear from students who are interested in pursuing their BSc, MSc or MD thesis in our field.
Please do not hesitate to contact us.
Translational proteomics
In translational proteomics, the Schilling laboratory pursues three major goals:
- establishing proteome profiles and markers to stratify patients with regard to therapeutic response and individualized therapeutic options
- establishing proteome profiles and markers that distinguish related diseases
- understanding mechanisms of therapy resistance and identifying novel therapeutic targets
We utilize three flavors of mass spectrometry based proteomics
- Explorative (“shotgun”)
- Targeted (MRM / SRM)
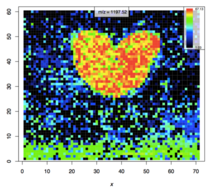
- Spatially resolved (“MALDI Imaging”, Fig. 1)
Proteolytic signaling
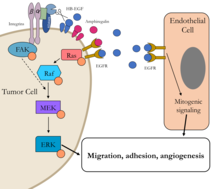
In this area, the laboratory focuses on cell surface proteases and how they modify cell-matrix interaction. Recent projects have investigated the roles of ADAM-9 and FAPα in solid tumors (Fig. 2).
Funding
The Schilling laboratory is supported, amongst others, by the Heisenberg program of the German Research Council (Deutsche Forschungsgemeinschaft, DFG), the CRC NephGen, the CRC OncoEscape, the RTG ProtPath, the RTG MeInBio, the DFG-FAPESP program, the DFG-ANR program, the ERA-Net programs on Personalized Medicine (PerMed) and Translational Cancer Research (TRANSCAN), the German Consortium for Translational Cancer Research (DKTK), the BMBF program “KMU-innovativ”, the invest BW program, and the Centers for Personalized Medicine Baden Württemberg.
Members of the Schilling laboratory
- Cabrita Figueiredo, Antonio Eduardo, MD (antonio.cabrita@uniklinik-freiburg.de)
- Dr. rer. nat. Fahrner, Matthias, Clinical Proteomics (matthias.fahrner@uniklinik-freiburg.de)
- Feilen, Tobias, Pharm. (tobias.feilen@uniklinik-freiburg.de)
- Fiestas Cueto, Ruth, cand. med. (ruth.fiestas.cueto@uniklinik-freiburg.de)
- Haberstroh, Hanna, M.Sc. (hanna.haberstroh@uniklinik-freiburg.de)
- Kirschstein, Raphael (raphael.kirschstein@uniklinik-freiburg.de)
- Li, Mujia, Pharm. (mujia.li@uniklinik-freiburg.de)
- Maldacker, Maximilian, M.Sc. (maximilian.maldacker@uniklinik-freiburg.de)
- Meyer, Larissa, M. Sc. (larissa.meyer@uniklinik-freiburg.de)
- Dr. rer. nat. Pinter, Niko, Proteogenomics (niko.pinter@uniklinik-freiburg.de)
- Seredynska, Adrianna, M.Sc. (adrianna.seredynska@uniklinik-freiburg.de)
- Thiery, Johanna, B.Sc. (johanna.thiery@uniklinik-freiburg.de)
- Dr. rer. nat. Tholen, Stefan, Proteomics Core Facility (stefan.tholen@uniklinik-freiburg.de)
- Schwaiger, Jessica, (jessica.schwaiger@uniklinik-freiburg.de)
- Topitsch, Annika, M.Sc. (annika.topitsch@uniklinik-freiburg.de)
- Vogele, Daniel, M.Sc. (daniel.vogele@uniklinik-freiburg.de)
- Weber, Stefanie (stefanie.weber@uniklinik-freiburg.de)
- Wehrle, Bettina, MTA (bettina.wehrle@uniklinik-freiburg.de)
- Werner, Tilman, Pharm. (tilman.werner@uniklinik-freiburg.de)
- Willeke, Christopher (christopher.willeke@uniklinik-freiburg.de)
Barta, Bálint András, MD
Bernhard, Patrick, M.Sc.
Dr. Campitelli de Barros, Bianca Carla Silva, M.Sc., Ph.D.
Dr. Chaves, Alison Felipe Alencar, M.Sc., Ph.D.
Dr. rer. nat. Chen, Chia-Yi
now Eurofins Biopharma
Cosenza Contreras, Miguel de Jesus, M.Sc.
Dinh,Thien-Ly Julia, M.Sc.
Dr. rer. nat. Fröhlich, Klemens
Proteomics Core Facility, University of Basel
Imberg, Fiona, cand. med. dent. Halstenbach, Tim, cand. med. dent.
Hause, Frank, M.Sc. M.A.
Goncharenko, Lina, B.Sc.
Dipl. Mol. Med. Knopf, Julia
now University of Heidelberg, laboratory of Prof. Dr. Marius Lemberg
Dr. rer. nat. Koczorowska, Maria
Lai, Zon Weng, PhD
now Harvard Medical School (Department of Cell Biology) and Harvard School of Public Health (Department of Genetics and Complex Diseases)
Langner, Anja Michelle, cand. med. dent.
Dr. rer. nat. Oria, Victor Oginga
Dept of Medical Oncology (Jilaveanu Lab), Yale Cancer Center, Yale University
Dr. rer. nat. Petrera, Agnese
now Helmholtz Zentrum München, Research Unit Protein Science
Dr. rer. nat. Sigloch, Florian
now PolyQuant GmbH, 93077 Bad Abbach, Germany
Dr. Stillger, Maren, M.Sc.
Dr. med. Werner, Janina, cand. med.
dr. med. Zirngibl, Max, MD
A data analysis approach for extracting information related to proteolytic activity (N-terminomics) from shotgun proteomics data). |
2023
- Mehta S, Bernt M, Chambers M, Fahrner M, Föll MC, Gruening B, Horro C, Johnson JE, Loux V, Rajczewski AT, Schilling O, Vandenbrouck Y, Gustafsson OJR, Thang WCM, Hyde C, Price G, Jagtap PD, Griffin TJ. A Galaxy of informatics resources for MS-based proteomics. Expert Rev Proteomics. 2023 Jul-Dec;20(11):251-266. doi: 10.1080/14789450.2023.2265062. Epub 2023 Oct 30. PMID: 37787106.
- Werner T, Fahrner M, Schilling O. Using proteomics for stratification and risk prediction in patients with solid tumors. Pathologie (Heidelb). 2023 Dec;44(Suppl 3):176-182. English. doi: 10.1007/s00292-023-01261-x. Epub 2023 Nov 24. PMID: 37999758.
- Rogg M, Maier JI, Helmstädter M, Sammarco A, Kliewe F, Kretz O, Weißer L, Van Wymersch C, Findeisen K, Koessinger AL, Tsoy O, Baumbach J, Grabbert M, Werner M, Huber TB, Endlich N, Schilling O, Schell C. A YAP/TAZ-ARHGAP29-RhoA Signaling Axis Regulates Podocyte Protrusions and Integrin Adhesions. Cells. 2023 Jul 6;12(13):1795. doi: 10.3390/cells12131795. PMID: 37443829; PMCID: PMC10340513.
- Cosenza-Contreras M, Schäfer A, Sing J, Cook L, Stillger MN, Chen CY, Hidalgo JV, Pinter N, Meyer L, Werner T, Bug D, Haberl Z, Kübeck O, Zhao K, Stei S, Gafencu AV, Ionita R, Brehar FM, Ferrer-Lozano J, Ribas G, Cerdá-Alberich L, Martí-Bonmatí L, Nimsky C, Van Straaten A, Biniossek ML, Föll M, Cabezas-Wallscheid N, Büscher J, Röst H, Arnoux A, Bartsch JW, Schilling O. Proteometabolomics of initial and recurrent glioblastoma highlights an increased immune cell signature with altered lipid metabolism. Neuro Oncol. 2023 Oct 26:noad208. doi: 10.1093/neuonc/noad208. Epub ahead of print. PMID: 37882631.
- Werner J, Bernhard P, Cosenza-Contreras M, Pinter N, Fahrner M, Pallavi P, Eberhard J, Bronsert P, Rückert F, Schilling O. Targeted and explorative profiling of kallikrein proteases and global proteome biology of pancreatic ductal adenocarcinoma, chronic pancreatitis, and normal pancreas highlights disease-specific proteome remodelling. Neoplasia. 2023 Feb;36:100871. doi: 10.1016/j.neo.2022.100871. Epub 2023 Jan 5. PMID: 36610378; PMCID: PMC9841175.
- Siegel PM, Barta BA, Orlean L, Steenbuck ID, Cosenza-Contreras M, Wengenmayer T, Trummer G, Wolf D, Westermann D, Schilling O, Diehl P. The serum proteome of VA-ECMO patients changes over time and allows differentiation of survivors and non-survivors: an observational study. J Transl Med. 2023 May 12;21(1):319. doi: 10.1186/s12967-023-04174-8. PMID: 37173738; PMCID: PMC10176307.
- Stillger MN, Chen CY, Lai ZW, Li M, Schäfer A, Pagenstecher A, Nimsky C, Bartsch JW, Schilling O. Changes in calpain-2 expression during glioblastoma progression predisposes tumor cells to temozolomide resistance by minimizing DNA damage and p53-dependent apoptosis. Cancer Cell Int. 2023 Mar 17;23(1):49. doi: 10.1186/s12935-023-02889-8. PMID: 36932402; PMCID: PMC10022304.
- Malik SC, Lin JD, Ziegler-Waldkirch S, Tholen S, Deshpande SS, Schwabenland M, Schilling O, Vlachos A, Meyer-Luehmann M, Schachtrup C. Tpr Misregulation in Hippocampal Neural Stem Cells in Mouse Models of Alzheimer's Disease. Cells. 2023 Dec 1;12(23):2757. doi: 10.3390/cells12232757. PMID: 38067185; PMCID: PMC10706632.
- Deichmann S, Schindel L, Braun R, Bolm L, Taylor M, Deshpande V, Schilling O, Bronsert P, Keck T, Ferrone C, Wellner U, Honselmann K. Overexpression of integrin alpha 2 (ITGA2) correlates with poor survival in patients with pancreatic ductal adenocarcinoma. J Clin Pathol. 2023 Aug;76(8):541-547. doi: 10.1136/jclinpath-2022-208176. Epub 2022 Apr 8. PMID: 35396216.
- Kocsmár É, Schmid M, Cosenza-Contreras M, Kocsmár I, Föll M, Krey L, Barta BA, Rácz G, Kiss A, Werner M, Schilling O, Lotz G, Bronsert P. Proteome alterations in human autopsy tissues in relation to time after death. Cell Mol Life Sci. 2023 Apr 5;80(5):117. doi: 10.1007/s00018-023-04754-3. PMID: 37020120; PMCID: PMC10075177.
- Thomsen AR, Sahlmann J, Bronsert P, Schilling O, Poensgen F, May AM, Timme-Bronsert S, Grosu AL, Vaupel P, Gebbers JO, Multhoff G, Lüchtenborg AM. Protocol of the HISTOTHERM study: assessing the response to hyperthermia and hypofractionated radiotherapy in recurrent breast cancer. Front Oncol. 2023 Dec 19;13:1275222. doi: 10.3389/fonc.2023.1275222. PMID: 38169879; PMCID: PMC10759986.
- Trevisan-Silva D, Cosenza-Contreras M, Oliveira UC, da Rós N, Andrade-Silva D, Menezes MC, Oliveira AK, Rosa JG, Sachetto ATA, Biniossek ML, Pinter N, Santoro ML, Nishiyama-Jr MY, Schilling O, Serrano SMT. Systemic toxicity of snake venom metalloproteinases: Multi-omics analyses of kidney and blood plasma disturbances in a mouse model. Int J Biol Macromol. 2023 Dec 31;253(Pt 6):127279. doi: 10.1016/j.ijbiomac.2023.127279. Epub 2023 Oct 6. PMID: 37806411.
- reprint (under revision): Miguel Cosenza-Contreras, Adrianna Seredynska, Niko Pinter, et al. Fragterminomics: extracting information on proteolytic processing from shotgun proteomics data processed by FragPipe. Authorea. November 04, 2023.
DOI: 10.22541/au.169906623.39670856/v1
- Dilza Trevisan-Silva, Miguel Cosenza-Contreras, Ursula C. Oliveira, Nancy da Rós, Débora Andrade-Silva, Milene C. Menezes, Ana Karina Oliveira, Jaqueline G. Rosa, Ana T.A. Sachetto, Martin L. Biniossek, Niko Pinter, Marcelo L. Santoro, Milton Y. Nishiyama-Jr, Oliver Schilling, Solange M.T. Serrano, Systemic toxicity of snake venom metalloproteinases: Multi-omics analyses of kidney and blood plasma disturbances in a mouse model, International Journal of Biological Macromolecules, Volume 253, Part 6, 2023, 127279, ISSN 0141-8130, doi.org/10.1016/j.ijbiomac.2023.127279
- Janina Werner, Patrick Bernhard, Miguel Cosenza-Contreras, Niko Pinter, Matthias Fahrner, Prama Pallavi, Johannes Eberhard, Peter Bronsert, Felix Rückert, Oliver Schilling, Targeted and explorative profiling of kallikrein proteases and global proteome biology of pancreatic ductal adenocarcinoma, chronic pancreatitis, and normal pancreas highlights disease-specific proteome remodelling, Neoplasia, Volume 36, 2023, 100871, ISSN 1476-5586, doi.org/10.1016/j.neo.2022.100871
- Thiery J, Fahrner M. Integration of proteomics in the molecular tumor board. Proteomics. 2023 Dec 24:e2300002. Epub ahead of print. PMID: 38143279. DOI: 10.1002/pmic.202300002
2022
-
Werner J, Bernhard P, Cosenza-Contreras M, Pinter N, Fahrner M, Pallavi P, Eberhard J, Bronsert P, Rückert F, Schilling O. Targeted and explorative profiling of kallikrein proteases and global proteome biology of pancreatic ductal adenocarcinoma, chronic pancreatitis, and normal pancreas highlights disease-specific proteome remodelling. Neoplasia. 2023 Jan 5;36:100871. doi: 10.1016/j.neo.2022.100871 . Epub ahead of print. PMID: 36610378.
-
Bernhard P, Feilen T, Rogg M, Fröhlich K, Cosenza-Contreras M, Hause F, Schell C, Schilling O. Proteome alterations during clonal isolation of established human pancreatic cancer cell lines. Cell Mol Life Sci. 2022 Oct 22;79(11):561. doi: 10.1007/s00018-022-04584-9 . PMID: 36271971 ; PMCID: PMC9587952.
-
Song K, Tholen S, Baumgartner D, Schilling O, Hess WR.
Isolation of intact and active FoF1 ATP synthase using a FLAG-tagged subunit from the cyanobacterium Synechocystis sp. PCC 6803.
STAR Protoc. 2022 Aug 16;3(3):101623. doi: 10.1016/j.xpro.2022.101623.
PMID: 36039073. -
Masilamani AP, Schulzki R, Yuan S, Haase IV, Kling E, Dewes F, Andrieux G, Börries M, Schnell O, Heiland DH, Schilling O, Ferrarese R, Carro MS. Calpain-mediated cleavage generates a ZBTB18 N-terminal product that regulates HIF1A signaling and glioblastoma metabolism.
iScience. 2022 Jun 17;25(7):104625. -
Budau KL, Sigel CS, Bergmann L, Lüchtenborg AM, Wellner U, Schilling O, Werner M, Tang L, Bronsert P.
Prognostic Impact of Tumor Budding in Intrahepatic Cholangiocellular Carcinoma.
J Cancer. 2022 May 6;13(8):2457-2471. -
Ruppert M, Barta BA, Korkmaz-Icöz S, Loganathan S, Oláh A, Sayour AA, Benke K, Nagy D, Bálint T, Karck M, Schilling O, Merkely B, Radovits T, Szabó G.
Sex similarities and differences in the reverse and anti-remodeling effect of pressure unloading therapy in a rat model of aortic banding and debanding.
Am J Physiol Heart Circ Physiol. 2022 Jul 1;323(1):H204-H222. -
Gonschorek P, Zorzi A, Maric T, Le Jeune M, Schüttel M, Montagnon M, Gómez-Ojea R, Vollmar DP, Whitfield C, Reymond L, Carle V, Verma H, Schilling O, Hovnanian A, Heinis C.
Phage Display Selected Cyclic Peptide Inhibitors of Kallikrein-Related Peptidases 5 and 7 and Their In Vivo Delivery to the Skin.
J Med Chem. 2022 Jun 2. doi: 10.1021/acs.jmedchem.2c00306. Online ahead of print. -
Bernhard P, Bretthauer BA, Brixius SJ, Bügener H, Groh JE, Scherer C, Damjanovic D, Haberstroh J, Trummer G, Benk C, Beyersdorf F, Schilling O, Pooth JS.
Serum proteome alterations during conventional and extracorporeal resuscitation in pigs.
J Transl Med. 2022 May 23;20(1):238. -
Fröhlich K, Brombacher E, Fahrner M, Vogele D, Kook L, Pinter N, Bronsert P, Timme-Bronsert S, Schmidt A, Bärenfaller K, Kreutz C, Schilling O. Benchmarking of analysis strategies for data-independent acquisition proteomics using a large-scale dataset comprising inter-patient heterogeneity.
Nat Commun. 2022 May 12;13(1):2622. -
Pinter N, Glätzer D, Fahrner M, Fröhlich K, Johnson J, Grüning BA, Warscheid B, Drepper F, Schilling O, Föll MC.
MaxQuant and MSstats in Galaxy Enable Reproducible Cloud-Based Analysis of Quantitative Proteomics Experiments for Everyone.
J Proteome Res. 2022 Jun 3;21(6):1558-1565. -
Fretwurst T, Tritschler I, Rothweiler R, Nahles S, Altmann B, Schilling O, Nelson K.Proteomic profiling of human bone from different anatomical sites - A pilot study.
Proteomics Clin Appl. 2022 Apr 24:e2100049. -
Rogg M, Maier JI, Ehle M, Sammarco A, Schilling O, Werner M, Schell C.
NUP133 Controls Nuclear Pore Assembly, Transcriptome Composition, and Cytoskeleton Regulation in Podocytes
Cells. 2022 Apr 7;11(8):1259. -
Föll MC, Volkmann V, Enderle-Ammour K, Timme S, Wilhelm K, Guo D, Vitek O, Bronsert P, Schilling O.
Moving translational mass spectrometry imaging towards transparent and reproducible data analyses: a case study of an urothelial cancer cohort analyzed in the Galaxy framework.
Clin Proteomics. 2022 Apr 19;19(1):8. -
Deichmann S, Schindel L, Braun R, Bolm L, Taylor M, Deshpande V, Schilling O, Bronsert P, Keck T, Ferrone C, Wellner U, Honselmann K.
Overexpression of integrin alpha 2 (ITGA2) correlates with poor survival in patients with pancreatic ductal adenocarcinoma.
J Clin Pathol. 2022 Apr 8:jclinpath-2022-208176. doi: 10.1136/jclinpath-2022-208176. Online ahead of print. -
Fahrner M, Föll MC, Grüning BA, Bernt M, Röst H, Schilling O.
Democratizing data-independent acquisition proteomics analysis on public cloud infrastructures via the Galaxy framework.
Gigascience. 2022 Feb 15. -
Kao WC, Ortmann de Percin Northumberland C, Cheng TC, Ortiz J, Durand A, von Loeffelholz O, Schilling O, Biniossek ML, Klaholz BP, Hunte C. Structural basis for safe and efficient energy conversion in a respiratory supercomplex
Nat Commun. 2022 Jan 27;13(1):545. -
Fayçal CA, Oszwald A, Feilen T, Cosenza-Contreras M, Schilling O, Loustau T, Steinbach F, Schachner H, Langer B, Heeringa P, Rees AJ, Orend G, Kain R.
An adapted passive model of anti-MPO dependent crescentic glomerulonephritis reveals matrix dysregulation and is amenable to modulation by CXCR4 inhibition.
Matrix Biol. 2022 Feb;106:12-33.
2021
- Alatibi KI, Tholen S, Wehbe Z, Hagenbuchner J, Karall D, Ausserlechner MJ, Schilling O, Grünert SC, Vockley J, Tucci S
Lipidomic and Proteomic Alterations Induced by Even and Odd Medium-Chain Fatty Acids on Fibroblasts of Long-Chain Fatty Acid Oxidation Disorders.
Int J Mol Sci. 2021 Sep 29;22(19):10556.
- Barta BA, Ruppert M, Fröhlich KE, Cosenza-Contreras M, Oláh A, Sayour AA, Kovács K, Karvaly GB, Biniossek M, Merkely B, Schilling O, Radovits T.
Sex-related differences of early cardiac functional and proteomic alterations in a rat model of myocardial ischemia.
J Transl Med. 2021 Dec 11;19(1):507.
-
Fahrner M, Bronsert P, Fichtner-Feigl S, Jud A, Schilling O.
Proteome biology of primary colorectal carcinoma and corresponding liver metastases.
Neoplasia. 2021 Dec;23(12):1240-1251. -
Erny D, Dokalis N, Mezö C, Castoldi A, Mossad O, Staszewski O, Frosch M, Villa M, Fuchs V, Mayer A, Neuber J, Sosat J, Tholen S, Schilling O, Vlachos A, Blank T, Gomez de Agüero M, Macpherson AJ, Pearce EJ, Prinz M.
Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease
Cell Metab. 2021 Nov 2;33(11):2260-2276. -
Barbieux C, Bonnet des Claustres M, Fahrner M, Petrova E, Tsoi LC, Gouin O, Leturcq F, Nicaise-Roland P, Bole C, Béziat V, Bourrat E, Schilling O, Gudjonsson JE, Hovnanian A.
Netherton syndrome subtypes share IL-17/IL-36 signature with distinct IFN-α and allergic responses.
J Allergy Clin Immunol. 2021 Sep 17:S0091-6749(21)01398-1.; online ahead of print. -
Braun R, Lapshyna O, Eckelmann S, Honselmann K, Bolm L, Ten Winkel M, Deichmann S, Schilling O, Kruse C, Keck T, Wellner U, Bronsert P, Brandenburger M.
Organotypic Slice Cultures as Preclinical Models of Tumor Microenvironment in Primary Pancreatic Cancer and Metastasis.
J Vis Exp. 2021 Jun 22;(172). -
Fahrner M, Kook L, Fröhlich K, Biniossek ML, Schilling O.
A Systematic Evaluation of Semispecific Peptide Search Parameter Enables Identification of Previously Undescribed N-Terminal Peptides and Conserved Proteolytic Processing in Cancer Cell Lines.
Proteomes. 2021 May 25;9(2):26. -
Klatt JN, Dinh TJ, Schilling O, Zengerle R, Schmidt F, Hutzenlaub T, Paust N.
Automation of peptide desalting for proteomic liquid chromatography - tandem mass spectrometry by centrifugal microfluidics.
Lab Chip. 2021 Jun 1;21(11):2255-2264. -
Maier JI, Rogg M, Helmstädter M, Sammarco A, Schilling O, Sabass B, Miner JH, Dengjel J, Walz G, Werner M, Huber TB, Schell C.
EPB41L5 controls podocyte extracellular matrix assembly by adhesome-dependent force transmission.
Cell Rep. 2021 Mar 23;34(12):108883. -
Eberhard J, Hirsch D, Schilling O, Dirks WG, Guo F, Fabarius A, Rückert F, Reißfelder C, Hohenberger P, Pallavi P.
First report on establishment and characterization of a carcinosarcoma tumour cell line model of the bladder.
Sci Rep. 2021 Mar 16;11(1):6030. -
Kunath A, Weiner J, Krause K, Rehders M, Pejkovska A, Gericke M, Biniossek ML, Dommel S, Kern M, Ribas-Latre A, Schilling O, Brix K, Stumvoll M, Klöting N, Heiker JT, Blüher M. Role of Kallikrein 7 in Body Weight and Fat Mass Regulation.
Biomedicines. 2021 Jan 29;9(2):131. -
Rogg M, Maier JI, Dotzauer R, Artelt N, Kretz O, Helmstädter M, Abed A, Sammarco A, Sigle A, Sellung D, Dinse P, Reiche K, Yasuda-Yamahara M, Biniossek ML, Walz G, Werner M, Endlich N, Schilling O, Huber TB, Schell C.
SRGAP1 Controls Small Rho GTPases To Regulate Podocyte Foot Process Maintenance.
J Am Soc Nephrol. 2021 Mar;32(3):563-579. -
Krauspe V, Fahrner M, Spät P, Steglich C, Frankenberg-Dinkel N, Maček B, Schilling O, Hess WR
Discovery of a small protein factor involved in the coordinated degradation of phycobilisomes in cyanobacteria.
Proc Natl Acad Sci U S A. 2021 Feb 2.
2020
-
Guo D, Föll MC, Volkmann V, Enderle-Ammour K, Bronsert P, Schilling O, Vitek O.
Deep multiple instance learning classifies subtissue locations in mass spectrometry images from tissue-level annotations.
Bioinformatics. 2020 Jul 1;36(Suppl_1):i300-i308. -
Honselmann KC, Finetti P, Birnbaum DJ, Monsalve CS, Wellner UF, Begg SKS, Nakagawa A, Hank T, Li A, Goldsworthy MA, Sharma H, Bertucci F, Birnbaum D, Tai E, Ligorio M, Ting DT, Schilling O, Biniossek ML, Bronsert P, Ferrone CR, Keck T, Mino-Kenudson M, Lillemoe KD, Warshaw AL, Fernández-Del Castillo C, Liss AS.
Neoplastic-Stromal Cell Cross-talk Regulates Matrisome Expression in Pancreatic Cancer.
Mol Cancer Res. 2020 Sep 1. -
Hoefflin R, Harlander S, Schäfer S, Metzger P, Kuo F, Schönenberger D, Adlesic M, Peighambari A, Seidel P, Chen CY, Consenza-Contreras M, Jud A, Lahrmann B, Grabe N, Heide D, Uhl FM, Chan TA, Duyster J, Zeiser R, Schell C, Heikenwalder M, Schilling O, Hakimi AA, Boerries M, Frew IJ.
HIF-1α and HIF-2α differently regulate tumour development and inflammation of clear cell renal cell carcinoma in mice.
Nat Commun. 2020 Aug 17;11(1):4111. -
Begg SKS, Birnbaum DJ, Clark JW, Mino-Kenudson M, Wellner UF, Schilling O, Lillemoe KD, Warshaw AL, Castillo CF, Liss AS.
FOLFIRINOX Versus Gemcitabine-based Therapy for Pancreatic Ductal Adenocarcinoma: Lessons from Patient-derived Cell Lines.
Anticancer Res. 2020 Jul;40(7):3659-3667. -
Gomez-Auli A, Hillebrand LE, Christen D, Günther SC, Biniossek ML, Peters C, Schilling O, Reinheckel T.
The secreted inhibitor of invasive cell growth CREG1 is negatively regulated by cathepsin proteases.
Cell Mol Life Sci. 2020 May 8. Online ahead of print. -
Shahinian JH, Rog-Zielinska EA, Schlimpert M, Mayer B, Tholen S, Kammerer B, Biniossek ML, Beyersdorf F, Schilling O, Siepe M.
Impact of left ventricular assist device therapy on the cardiac proteome and metabolome composition in ischemic cardiomyopathy.
Artif Organs. 2020 Mar;44(3):257-267. -
Baumert HM, Metzger E, Fahrner M, George J, Thomas RK, Schilling O, Schüle R
Depletion of histone methyltransferase KMT9 inhibits lung cancer cell proliferation by inducing non-apoptotic cell death.
Cancer Cell Int. 2020 Feb 17;20:52. -
Capponi S, Stöffler N, Irimia M, Van Schaik FMA, Ondik MM, Biniossek ML, Lehmann L, Mitschke J, Vermunt MW, Creyghton MP, Graybiel AM, Reinheckel T, Schilling O, Blencowe BJ, Crittenden JR, Timmers HTM.
Neuronal-specific microexon splicing of TAF1 mRNA is directly regulated by SRRM4/nSR100.
RNA Biol. 2020 Jan;17(1):62-74.
2019
- Föll MC, Moritz L, Wollmann T, Stillger MN, Vockert N, Werner M, Bronsert P, Rohr K, Grüning BA, Schilling O.
Accessible and reproducible mass spectrometry imaging data analysis in Galaxy.
Gigascience. 2019 Dec 1;8(12).
- Behringer S, Wingert V, Oria V, Schumann A, Grünert S, Cieslar-Pobuda A, Kölker S, Lederer AK, Jacobsen DW, Staerk J, Schilling O, Spiekerkoetter U, Hannibal, L. Targeted Metabolic Profiling of Methionine Cycle Metabolites and Redox Thiol Pools in Mammalian Plasma, Cells and Urine.
Metabolites. 2019 Oct 18;9(10).
- Larionov A, Dahlke E, Kunke M, Zanon Rodriguez L, Schiessl IM, Magnin JL, Kern U, Alli AA, Mollet G, Schilling O, Castrop H, Theilig F.
Cathepsin B increases ENaC activity leading to hypertension early in nephrotic syndrome.
J Cell Mol Med. 2019 Oct;23(10):6543-6553.
- Rigogliuso G, Biniossek ML, Goodier JL, Mayer B, Pereira GC, Schilling O, Meese E, Mayer J.
A human endogenous retrovirus encoded protease potentially cleaves numerous cellular proteins.
Mob DNA. 2019 Aug 22;10:36.
- Weise SC, Arumugam G, Villarreal A, Videm P, Heidrich S, Nebel N, Dumit VI, Sananbenesi F, Reimann V, Craske M, Schilling O, Hess WR, Fischer A, Backofen R, Vogel T.
FOXG1 Regulates PRKAR2B Transcriptionally and Posttranscriptionally via miR200 in the Adult Hippocampus.
Mol Neurobiol. 2019 Jul;56(7):5188-5201.
- Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, Ebright RY, Karabacak MN, Kulkarni AS, Liu A, Vincent Jordan N, Franses JW, Philipp J, Kreuzer J, Desai N, Arora KS, Rajurkar M, Horwitz E, Neyaz A, Tai E, Magnus NKC, Vo KD, Yashaswini CN, Marangoni F, Boukhali M, Fatherree JP, Damon LJ, Xega K, Desai R, Choz M, Bersani F, Langenbucher A, Thapar V, Morris R, Wellner UF, Schilling O, Lawrence MS, Liss AS, Rivera MN, Deshpande V, Benes CH, Maheswaran S, Haber DA, Fernandez-Del-Castillo C, Ferrone CR, Haas W, Aryee MJ, Ting DT.
Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer.
Cell. 2019 Jun 27;178(1):160-175.
- Li M, Müller C, Fröhlich K, Gorka O, Zhang L, Groß O, Schilling O, Einsle O, Jessen-Trefzer C.
Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol. 2019 Jun 20;26(6):852-862.e6.
- Zamboglou C, Carles M, Fechter T, Kiefer S, Reichel K, Fassbender TF, Bronsert P, Koeber G, Schilling O, Ruf J, Werner M, Jilg CA, Baltas D, Mix M, Grosu AL.
Radiomic features from PSMA PET for non-invasive intraprostatic tumor discrimination and characterization in patients with intermediate- and high-risk prostate cancer - a comparison study with histology reference.
Theranostics. 2019 Apr 13;9(9):2595-2605.
- Ohmer M, Tzivelekidis T, Niedenführ N, Volceanov-Hahn L, Barth S, Vier J, Börries M, Busch H, Kook L, Biniossek ML, Schilling O, Kirschnek S, Häcker G.
Infection of HeLa cells with Chlamydia trachomatis inhibits protein synthesis and causes multiple changes to host cell pathways.
Cell Microbiol. 2019 Apr;21(4):e12993.
- Siemer S, Westmeier D, Barz M, Eckrich J, Wünsch D, Seckert C, Thyssen C, Schilling O, Hasenberg M, Pang C, Docter D, Knauer SK, Stauber RH, Strieth S.
Biomolecule-corona formation confers resistance of bacteria to nanoparticle-induced killing: Implications for the design of improved nanoantibiotics.
Biomaterials. 2019 Feb;192:551-559.
- Oria VO, Lopatta P, Schmitz T, Preca BT, Nyström A, Conrad C, Bartsch JW, Kulemann B, Hoeppner J, Maurer J, Bronsert P, Schilling O.
ADAM9 contributes to vascular invasion in pancreatic ductal adenocarcinoma.
Mol Oncol. 2019 Feb;13(2):456-479.
- Zhang HE, Hamson EJ, Koczorowska MM, Tholen S, Chowdhury S, Bailey CG, Lay AJ, Twigg SM, Lee Q, Roediger B, Biniossek ML, O'Rourke MB, McCaughan GW, Keane FM, Schilling O, Gorrell MD.
Identification of Novel Natural Substrates of Fibroblast Activation Protein-alpha by Differential Degradomics and Proteomics.
Mol Cell Proteomics. 2019 Jan;18(1):65-85.
2018
- Oria VO, Lopatta P, Schmitz T, Preca BT, Nyström A, Conrad C, Bartsch JW, Kulemann B, Hoeppner J, Maurer J, Bronsert P, Schilling O.
ADAM9 contributes to vascular invasion in pancreatic ductal adenocarcinoma.
Mol Oncol. 2018 Dec 17. doi: 10.1002/1878-0261.12426.
Pubmed 30556643 [Epub ahead of print].
- Ohmer M, Tzivelekidis T, Niedenführ N, Volceanov-Hahn L, Barth S, Vier J, Börries M, Busch H, Kook L, Biniossek ML, Schilling O, Kirschnek S, Häcker G.
Infection of HeLa cells with Chlamydia trachomatis inhibits protein synthesis and causes multiple changes to host cell pathways.
Cell Microbiol. 2018 Dec 14:e12993. doi: 10.1111/cmi.12993.
Pubmed 30551267 [Epub ahead of print].
- Weise SC, Arumugam G, Villarreal A, Videm P, Heidrich S, Nebel N, Dumit VI, Sananbenesi F, Reimann V, Craske M, Schilling O, Hess WR, Fischer A, Backofen R, Vogel T.FOXG1 Regulates PRKAR2B Transcriptionally and Posttranscriptionally via miR200 in the Adult Hippocampus.
Mol Neurobiol. 2018 Dec 11. doi: 10.1007/s12035-018-1444-7.
Pubmed 30539330 [Epub ahead of print].
- Enderle-Ammour K, Wellner U, Kocsmar E, Kiss A, Lotz G, Csanadi A, Bader M, Schilling O, Werner M, Bronsert P.
[Three-dimensional reconstruction of solid tumors : Morphological evidence for tumor heterogeneity].
Pathologe. 2018 Dec;39(Suppl 2):231-235. doi: 10.1007/s00292-018-0529-4.
Pubmed 30361775 Review.
- Wang P, Magdolen V, Seidl C, Dorn J, Drecoll E, Kotzsch M, Yang F, Schmitt M, Schilling O, Rockstroh A, Clements JA, Loessner D.
Kallikrein-related peptidases 4, 5, 6 and 7 regulate tumour-associated factors in serous ovarian cancer.
Br J Cancer. 2018 Oct;119(7):823-831. doi: 10.1038/s41416-018-0260-1. Epub 2018 Oct 5. Pubmed 30287916.
- Oria VO, Bronsert P, Thomsen AR, Föll MC, Zamboglou C, Hannibal L, Behringer S, Biniossek ML, Schreiber C, Grosu AL, Bolm L, Rades D, Keck T, Werner M, Wellner UF, Schilling O. Proteome Profiling of Primary Pancreatic Ductal Adenocarcinomas Undergoing Additive Chemoradiation Link ALDH1A1 to Early Local Recurrence and Chemoradiation Resistance.
Transl Oncol. 2018 Dec;11(6):1307-1322. doi: 10.1016/j.tranon.2018.08.001. Epub 2018 Aug 30.
Pubmed 30172883.
- Porodko A, Cirnski A, Petrov D, Raab T, Paireder M, Mayer B, Maresch D, Nika L, Biniossek ML, Gallois P, Schilling O, Oostenbrink C, Novinec M, Mach L.
The two cathepsin B-like proteases of Arabidopsis thaliana are closely related enzymes with discrete endopeptidase and carboxydipeptidase activities.
Biol Chem. 2018 Sep 25;399(10):1223-1235. doi: 10.1515/hsz-2018-0186.
Pubmed 29924726.
- Haun F, Neumann S, Peintner L, Wieland K, Habicht J, Schwan C, Østevold K, Koczorowska MM, Biniossek M, Kist M, Busch H, Boerries M, Davis RJ, Maurer U, Schilling O, Aktories K, Borner C.
Identification of a novel anoikis signalling pathway using the fungal virulence factor gliotoxin. Nat Commun. 2018 Aug 30;9(1):3524. doi: 10.1038/s41467-018-05850-w.
Pubmed 30166526.
- Schilling O, Biniossek ML, Mayer B, Elsässer B, Brandstetter H, Goettig P, Stenman UH, Koistinen H.
Specificity profiling of human trypsin-isoenzymes.
Biol Chem. 2018 Sep 25;399(9):997-1007. doi: 10.1515/hsz-2018-0107.
Pubmed 29883318.
- Chen CY, Melo E, Jakob P, Friedlein A, Elsässer B, Goettig P, Kueppers V, Delobel F, Stucki C, Dunkley T, Fauser S, Schilling O, Iacone R.
N-Terminomics identifies HtrA1 cleavage of thrombospondin-1 with generation of a proangiogenic fragment in the polarized retinal pigment epithelial cell model of age-related macular degeneration.
Matrix Biol. 2018 Sep;70:84-101. doi: 10.1016/j.matbio.2018.03.013. Epub 2018 Mar 20.
Pubmed29572155.
- Drendel V, Heckelmann B, Schell C, Kook L, Biniossek ML, Werner M, Jilg CA, Schilling O. Proteomic distinction of renal oncocytomas and chromophobe renal cell carcinomas.
Clin Proteomics. 2018 Aug 3;15:25. doi: 10.1186/s12014-018-9200-6. eCollection 2018. Pubmed 30087584.
- Debela M, Magdolen V, Skala W, Elsässer B, Schneider EL, Craik CS, Biniossek ML, Schilling O, Bode W, Brandstetter H, Goettig P.
Structural determinants of specificity and regulation of activity in the allosteric loop network of human KLK8/neuropsin.
Sci Rep. 2018 Jul 16;8(1):10705. doi: 10.1038/s41598-018-29058-6.
Pubmed 30013126.
- Westmeier D, Solouk-Saran D, Vallet C, Siemer S, Docter D, Götz H, Männ L, Hasenberg A, Hahlbrock A, Erler K, Reinhardt C, Schilling O, Becker S, Gunzer M, Hasenberg M, Knauer SK, Stauber RH.
Nanoparticle decoration impacts airborne fungal pathobiology.
Proc Natl Acad Sci U S A. 2018 Jul 3;115(27):7087-7092. doi: 10.1073/pnas.1804542115 Pubmed 29925597.
- Oria VO, Lopatta P, Schilling O.
The pleiotropic roles of ADAM9 in the biology of solid tumors.
Cell Mol Life Sci. 2018 Jul;75(13):2291-2301. doi: 10.1007/s00018-018-2796-x. Epub 2018 Mar 17.
Pubmed29550974 Review.
- Seidl M, Bader M, Vaihinger A, Wellner UF, Todorova R, Herde B, Schrenk K, Maurer J, Schilling O, Erbes T, Fisch P, Pfeiffer J, Hoffmann L, Franke K, Werner M, Bronsert P. Morphology of Immunomodulation in Breast Cancer Tumor Draining Lymph Nodes Depends on Stage and Intrinsic Subtype.
Sci Rep. 2018 Mar 28;8(1):5321. doi: 10.1038/s41598-018-23629-3.
Pubmed 29593307.
- Föll MC, Fahrner M, Oria VO, Kühs M, Biniossek ML, Werner M, Bronsert P, Schilling O.
Reproducible proteomics sample preparation for single FFPE tissue slices using acid-labile surfactant and direct trypsinization.
Clin Proteomics. 2018 Mar 6;15:11. doi: 10.1186/s12014-018-9188-y. eCollection 2018. Pubmed 29527141.
- Föll MC, Fahrner M, Gretzmeier C, Thoma K, Biniossek ML, Kiritsi D, Meiss F, Schilling O, Nyström A, Kern JS.
Identification of tissue damage, extracellular matrix remodeling and bacterial challenge as common mechanisms associated with high-risk cutaneous squamous cell carcinomas.
Matrix Biol. 2018 Mar;66:1-21. doi: 10.1016/j.matbio.2017.11.004. Epub 2017 Nov 20. Pubmed 29158163.
- Müller AK, Föll M, Heckelmann B, Kiefer S, Werner M, Schilling O, Biniossek ML, Jilg CA, Drendel V.
Proteomic Characterization of Prostate Cancer to Distinguish Nonmetastasizing and Metastasizing Primary Tumors and Lymph Node Metastases.
Neoplasia. 2018 Feb;20(2):140-151. doi: 10.1016/j.neo.2017.10.009. Epub 2017 Dec 14. Pubmed 29248718.
- Melo E, Oertle P, Trepp C, Meistermann H, Burgoyne T, Sborgi L, Cabrera AC, Chen CY, Hoflack JC, Kam-Thong T, Schmucki R, Badi L, Flint N, Ghiani ZE, Delobel F, Stucki C, Gromo G, Einhaus A, Hornsperger B, Golling S, Siebourg-Polster J, Gerber F, Bohrmann B, Futter C, Dunkley T, Hiller S, Schilling O, Enzmann V, Fauser S, Plodinec M, Iacone R.
HtrA1 Mediated Intracellular Effects on Tubulin Using a Polarized RPE Disease Model.
EBioMedicine. 2018 Jan;27:258-274. doi: 10.1016/j.ebiom.2017.12.011. Epub 2017 Dec 13. Pubmed 29269042.
Kontakt Forschung
Generelle Anfragen zur Forschung am Institut
pathologie.projektkoord@uniklinik-freiburg.de
Forschungsverbünde:
- CCCF
- CCI
- DKTK
- CRC 1453 Nephrogenetics
- CRC 1479 OncoEscape
- CRC 1160 IMPATH
- RTG 2606 ProPath
- CRU 329 podocyte
- Genom DE
- RTG 2344 MeinBioInMe
- nNGM Lung Cancer
- ZPM
- dnpm
- ERA Transcan projects (PREDICO, ICC-STRAT)
- HORUS
- Memorial Sloan Kettering Cancer Center collaboration
Institut für Klinische Pathologie
Universitätsklinikum Freiburg
Breisacher Str. 115a
79106 Freiburg
Tel.: +49 761 270 80060 (Sekretariat)
Fax: +49 761 270-80040
pathologie.direktion@uniklinik-freiburg.de
Postanschrift:
Postfach 214
79002 Freiburg