Duyster Laboratory
Scientific focus
"Niche hijacking“ by leukemic cells has been shown to contribute to leukemic cell expansion as well as enhanced survival of leukemic stem cells. However, the factors that mediate this are mainly unknown so far.
We found the STAT5 target gene Oncostatin M (OSM) overexpressed in certain mouse models of acute myeloid leukemia (AML) and myeloproliferative syndromes (MPN). We and others could show, that OSM overexpression in a murine bone marrow (BM) transplantation model leads to the induction of a lethal myeloproliferative disease. The OSM receptor OSMR however, is not expressed on leukemia cells or hematopoietic cells in general, but can be found on BM niche cells. In preliminary studies, we have analyzed the effects of OSM on these cells in vitro. We could demonstrate increased cytokine production and metabolic activity after OSM treatment. Furthermore, we found that murine models of OSM overexpression show markedly increased numbers of exhausted T cells and upregulation of immune inhibitory cell surface molecules. Thus, activation of the OSMR on niche cells may lead to an inflammatory environment, which favors leukemic expansion and promote immune escape.
To test this hypothesis, we study T cell function and leukemia induction in OSMR and OSM knockout leukemia mice models. We address the effects of OSM on BM stroma cells in more detail and try to replicate the in vivo effects of OSM by co-culture of T cells in combination with leukemic and stroma cells. In order to validate the clinical impact of these findings, we analyze serum and leukemic cells from patients of different disease entities for OSM expression.
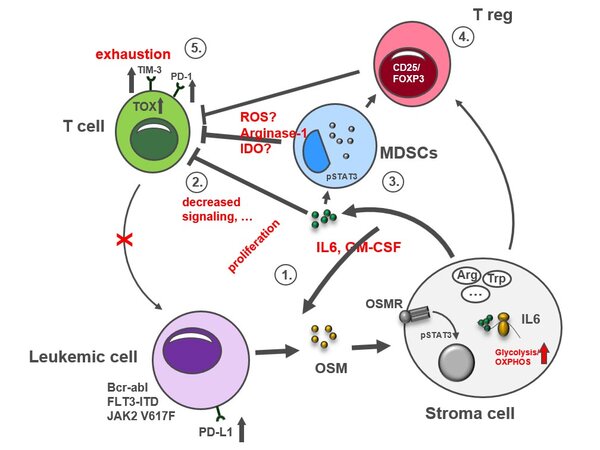
Hypothetical model of OSM signaling in leukemic diseases
The IL-6-faimily cytokine OSM is produced by leukemic cells harboring STAT5–activating mutations and released into the BM microenvironment. In a paracrine manner, BM niche cells respond to OSM and induce a immunosuppressive microenvironment promoting immune escape and leukemic cell expansion.
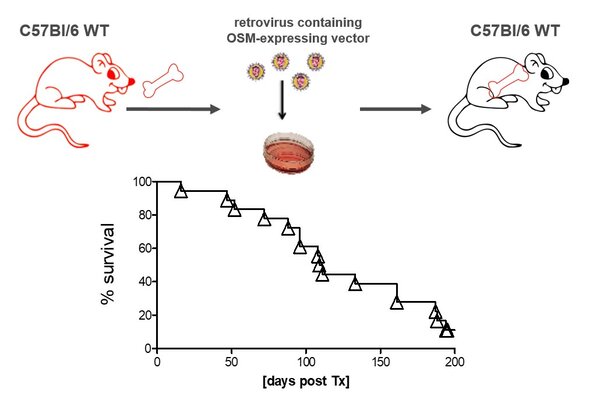
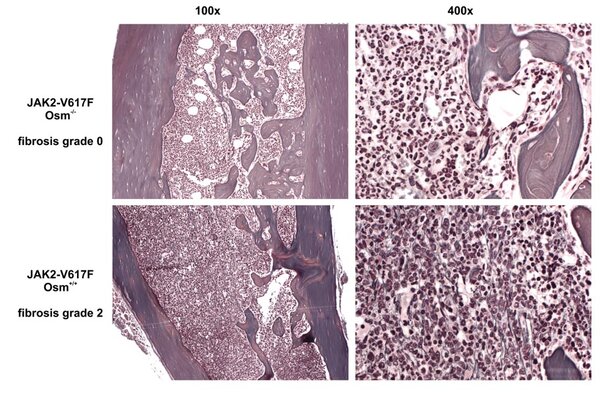
OSM expression mouse model
OSM expressing animal model.
Mice transplanted with BM retrovirally transfected with an OSM-expressing vector develop a deadly MPN-like disease.
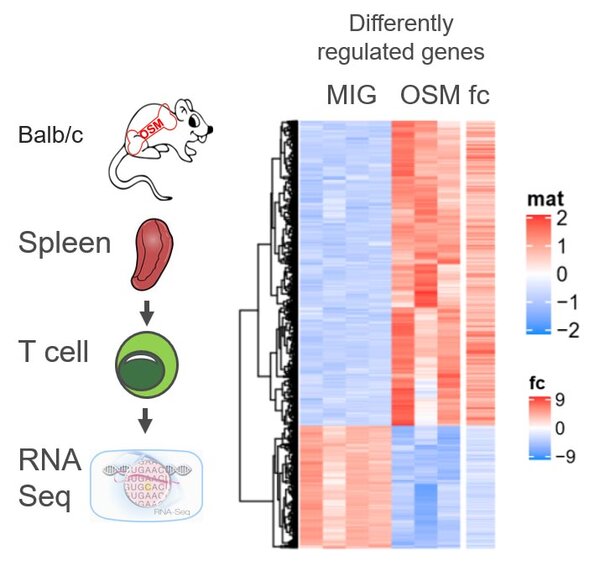
RNA Seq splenic T cells
RNA-seq of isolated T cells from OSM expressing mic.
RNA sequencing of RNA isolated from splenic T cells from OSM expressing Balb/c mice show substantially differently regulated genes.
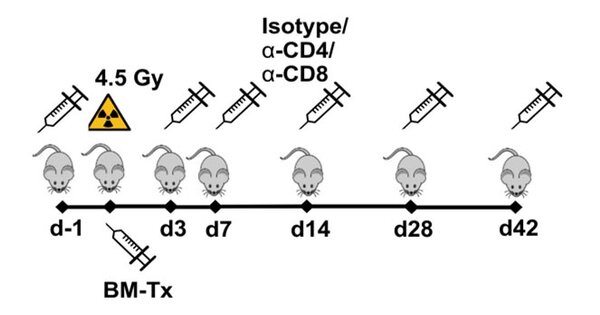
Immune cell depletion
Treatment schedule for the injection of T cell depletion antibodies targeting CD4 or CD8.
To delineate the role of OSM on the adapt and acquired immune system in mice transplanted with BM (OSM+/+ vs. OSM-/-) transfected with STAT5-activating oncogenic mutations, mice are treated with immune cell depleting antibodies, e.g. targeting CD4+- or CD8+-T-cells.
Team
Dr. med. Michael Rassner
post-doc
Kirstyn Anne Crossley
PhD candidate
Kornelia Fritsch
Med. Techn. Assistentin
Further Information
Dr. Siva Hari Gorantla
post-doc
Dr. Tony Müller
post-doc
- DFG: The Hematopoietic Niches FOR 2033
- José Carreras scholarship DJCLS 02FN/2017
- Leticia Quintanilla-Martinez, Department of Pathology and Neuropathology, University Hospital Tübingen & Comprehensive Cancer Center Tübingen
- Gerhard Müller-Newen, RWTH Aachen
- Robert Oostendorp, Klinikum rechts der Isar, TU München
- Claudia Waskow, Medical Faculty Carl Gustav Carus, TU Dresden
- Melanie Börries, Institut für Medizinische Bioinformatik und Systemmedizin, Freiburg
- Robert Zeiser, Klinik für Innere Medizin I, Tumorimmunologie und Immunregulation, Uniklinik Freiburg
- Thomas Reinheckel, Institut für Medizinische Bioinformatik und Systemmedizin, Freiburg
- Tilman Brummer, Institut für Medizinische Bioinformatik und Systemmedizin, Freiburg
- Olaf Groß, Institute of Neuropathology, Neurozentrum, University Medical Center Freiburg
- Cabezas-Wallscheid, Max-Planck-Institut für Immunbiologie und Epigenetik, Freiburg
- Lena Illert, Klinik für Innere Medizin I, Tumorzentrum - CCCF, Uniklinik Freiburg
- Fetsch V, …, Duyster J, Börries M, Hofmann M, Färber J, Braun LM, Zähringer A, Lübbert M, Toffalori C, Vago L, Heidel FH, Minguet S, Apostolova P, Feuchtinger T, Maas-Bauer K, Blaeschke F, Kühn MWM, Timmers M, Wertheimer T, Perner F, Zeiser R. Menin inhibition enhances graft-versus-leukemia effects by T-cell activation and endogenous retrovirus induction in AML. Blood. 2025 Oct 27:blood.2025029712. doi: 10.1182/blood.2025029712.
- Zwick M, …, Duyster J, Schetelig J, Brummer T, Zeiser R, Köhler N. FLT3-ITD Induces CMTM6 and Enhances Immune Escape in Acute Myeloid Leukemia. Cancer Res. 2025 Oct 3. doi: 10.1158/0008-5472.CAN-25-0349.
- Khan R, Phely L, Ehrenfeld S, Schmitz T, Veratti P, Wolfes J, Shoumariyeh K, Andrieux G, Martens US, Bra S, Auer M, Schilling O, Boerries M, Speicher M, Illert AL, Duyster J, Miething C. Modeling the t(2;5) Translocation of Anaplastic Large Cell Lymphoma Using CRISPR-Mediated Chromosomal Engineering. Cancers (Basel). 2025 Jul 2;17(13):2226. doi: 10.3390/cancers17132226.
- Gorantla SP, Rassner M, Crossley KA, Müller TA, Poggio T, Khaja Saleem S, Kleinfelder H, Gambheer SMM, Endres C, Schaberg S, Schmidt D, Prince G, Gonzalez-Menendez I, Bentrop D, Trittler R, Rylova S, Pfeifer D, Andrieux G, Quintanilla-Martinez L, Illert AL, von Bubnoff N, Zeiser R, Duyster J. Efficacy of JAK1/2 inhibition in murine myeloproliferative neoplasms is not mediated by targeting oncogenic signaling. Nat Commun. 2025 May 24;16(1):4833. doi: 10.1038/s41467-025-60019-6.
- Gorantla SP, Oelschläger L, Prince G, Osius J, Kolluri SB, Maluje Y, Fähnrich A, Ernst N, Barbosa Gulde A, Ludwig RJ, Gemoll T, Fliedner S, Walter W, Haferlach T, Gebauer N, Busch H, Duyster J, von Bubnoff N. Ruxolitinib mediated paradoxical JAK2 hyperphosphorylation is due to the protection of activation loop tyrosines from phosphatases. Leukemia. 2025 Jul;39(7):1678-1691. doi: 10.1038/s41375-025-02594-7. Epub 2025 Apr 23.
- Kuehn JC, Neidert NN, Zhang J, Mutter J, Alig S, Klingler C, Hummel F, Ranganathan L, Bleul S, Beck J, Prinz M, Diehn M, Alizadeh A, Duyster J, Sankowski R, Heiland DH, Scherer F. The effect of ibrutinib on the myeloid cell compartment in CNS lymphoma. Leukemia. 2025 Jun;39(6):1532-1535. doi: 10.1038/s41375-025-02600-y. Epub 2025 Apr 10.
- Kusterer M, Lahnalampi M, Voutilainen M, Brand A, Pennisi S, Norona J, Gentile G, Herzog H, Greve G, Lübbert M, Sipola M, Kaartinen E, Sankowski R, Prinz M, Killmer S, Lago MS, Bengsch B, Cysar SR, Aumann K, Werner M, Duyster J, Lohi O, Heinäniemi M, Duque-Afonso J. Dynamic evolution of TCF3-PBX1 leukemias at the single-cell level under chemotherapy pressure. Hemasphere. 2025 Feb 3;9(2):e70071. doi: 10.1002/hem3.70071. eCollection 2025 Feb.
- Shoumariyeh K, Csernalabics B, Salimi Alizei E, Reinscheid M, Giese S, Ciminski K, Kochs G, Schwemmle M, Lang-Meli J, Maas M, Roehlen N, Karl V, Graeser A, Sogukpinar O, von Metzler I, Grathwohl D, Rasche L, Hebart H, Kull M, Emmerich F, Waller CF, Duyster J, Engelhardt M, Hartmann TN, Bengsch B, Boettler T, Neumann-Haefelin C, Hofmann M, Thimme R, Luxenburger H. Impaired SARS-CoV-2-Specific CD8+ T Cells After Infection or Vaccination but Robust Hybrid T Cell Immunity in Patients with Multiple Myeloma. Vaccines (Basel). 2024 Nov 1;12(11):1249. doi: 10.3390/vaccines12111249.
- Kreutmair S, …, Duyster J, Illert AL: Loss of the Fanconi anemia-associated protein NIPA causes bone marrow failure. 2020, J Clin Invest; 130:2827-2844.
- Kreutmair S, …, Duyster J, Illert AL: Existence of reprogrammed lymphoma stem cells in a murine ALCL-like model. 2020, Leukemia; 17: doi: 10.1038/s41375-020-0789-x
- Waldeck S, Rassner M, …, Duyster J, von Bubnoff N. CCL5 mediates target-kinase independent resistance to FLT3 inhibitors in FLT3-ITD-positive AML. 2020, Mol Oncol; 14:779-794.
- Phosphorylation of BECLIN-1 by BCR-ABL suppresses autophagy in chronic myeloid leukemia. Yu C, Gorantla SP, … Müller TA, … Duyster J, Illert AL. 2020, Haematologica; 105:1285-1293.
- NPM1c alters FLT3-D835Y localization and signaling in acute myeloid leukemia. Rudorf A, Müller TA, … Gorantla SP, …Duyster J, Illert AL. 2020, Blood; 134:383-388.
- Jilg S, Rassner M, … Duyster J, ..von Bubnoff N. Circulating cKIT and PDGFRA DNA indicates disease activity in Gastrointestinal Stromal Tumor (GIST). 2019, Int J Cancer; 2292-2303.
- Müller TA, …, Duyster J. PIM1 inhibition effectively enhances plerixafor-induced HSC mobilization by counteracting CXCR4 upregulation and blocking CXCL12 secretion. 2019, Leukemia; 33:1296-1301.
- Dr. M. Rassner - Young Investigator award DGHO 2020
- Dr. M. Rassner - Swiss GIST prize
- Dr. M. Rassner - Young Investigator award DGHO 2018
- Dr. M. Rassner - Poster prize DGHO 2017