Hoefflin Laboratory
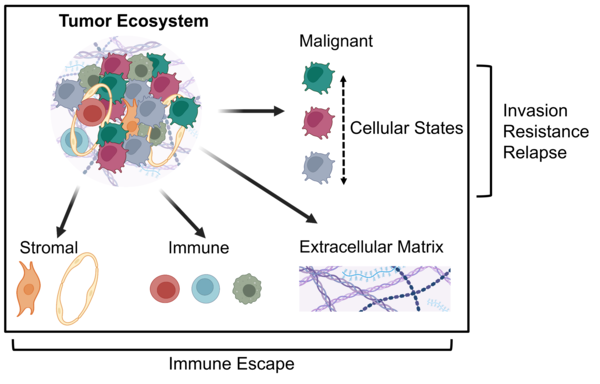
About usThe Hoefflin Lab combines cutting-edge experimental and computational approaches to decode the tumor ecosystem in time and space at single-cell resolution.
We leverage spatial multi-omics technologies and advanced computation and focus on how cancer evades the immune system and infiltrates surrounding healthy tissue.
These two hallmarks—immune escape and invasive behavior—make cancer notoriously difficult to treat. Our overarching goal is to overcome these challenges by developing new therapeutic concepts.

Immune escape and invasion in glioma
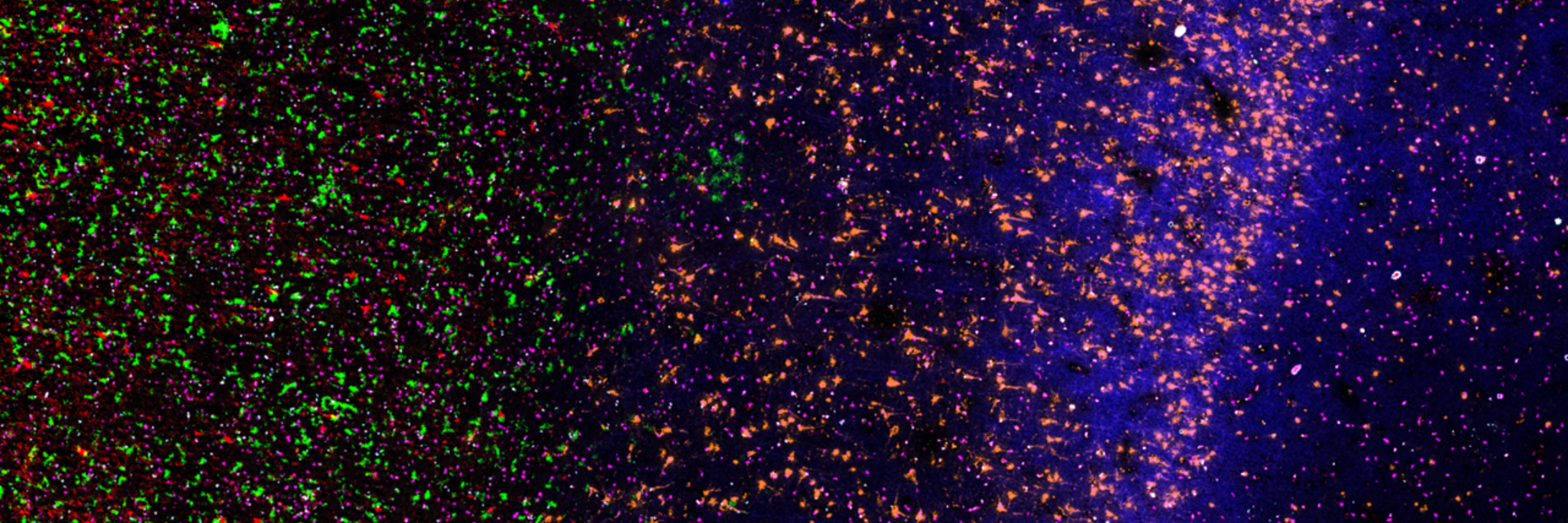
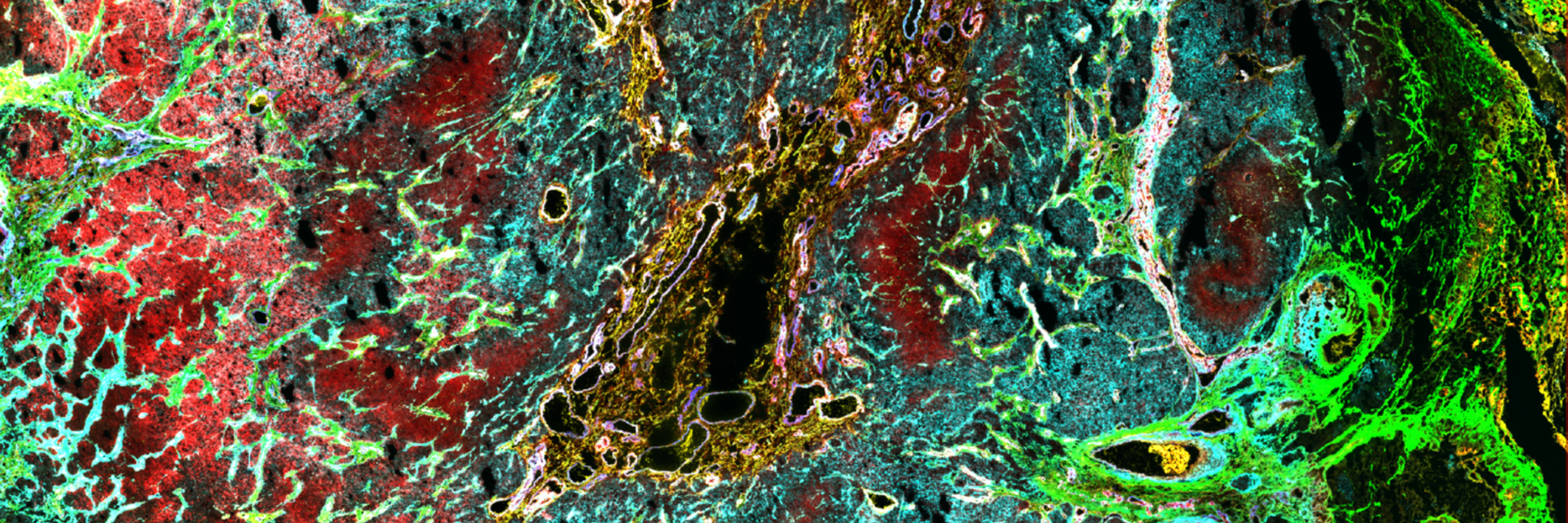
One focus of our lab is to decipher the interplay between cancer cell states, the extracellular matrix (ECM) they produce, and the associated mechanisms of immune evasion and invasive behavior in gliomas. We approach this by integrating novel spatial single-cell methods at both the proteomic and transcriptomic levels, using advanced computational approaches to precisely characterize these interactions (Nature 2024, Cell 2024). Our work includes profiling human glioma samples, as well as ex vivo organoid and in vivo mouse models. The ultimate goal is to develop new therapeutic strategies, including those that target the ECM.
Drivers of spatial organization in cancer
We have previously identified cancer cell states that consistently recur across tumor entities, suggesting they represent core programs of cancer (Nature Cancer 2025, Nature 2023). While these states are partly driven by intrinsic factors such as genetic mutations, they are predominantly shaped by extrinsic cues from the tumor microenvironment—including neighboring cells, soluble factors like ligands and cytokines, and components of the extracellular matrix (ECM). Our aim is to dissect these cell–cell and cell–ECM interactions using spatial proteomic and transcriptomic technologies across a range of human and murine cancer types. Ultimately, we seek to identify the external drivers of specific cancer cell states and leverage this knowledge to develop cell state–targeted therapies.
Computational methods development for spatial analysis
The analysis of spatial omics data is an emerging field with few established standards. In particular, spatial proteomics requires a labor-intensive, iterative workflow that involves interpreting multiplexed imaging data and analyzing the resulting protein expression matrices obtained through segmentation (Nature 2024, Cell 2024). These steps are typically decoupled, as they rely on separate software tools—image viewers for visualization and statistical programming environments for analysis. Our work focuses on seamlessly integrating these processes by developing user-friendly workflows and tools that enable interactive spatial visualization and analysis of millions of cells.
Team

Principal Investigator
Head of Hoefflin Laboratory
Dr. Rouven Höfflin
rouven.hoefflin@uniklinik-freiburg.de
Zentrum Translationale Zellforschung (ZTZ)
Breisacher Str. 115
D- 79106 Freiburg
📘 Google Scholar
🆔 ORCID
✖ X


Lingling Zhang, PhD
Postdoc und Visiting Scientist

Open Positions

We welcome initiative applications from PhD, MD, and Master's students who are passionate about exploring the complexity of the tumor ecosystem. We offer dry-lab, wet-lab, and hybrid projects that bridge experimental and computational approaches. For PhD positions, we particularly encourage applicants with backgrounds in molecular biology, bioinformatics, computer science, or related fields. Our lab provides a highly interdisciplinary environment, combining cutting-edge spatial technologies with functional experimental strategies. If you're excited to contribute to ongoing projects—or to develop your own ideas within our research focus—we would be delighted to hear from you.
International:
- Mario Suva (Harvard, Boston, USA)
- Itay Tirosh (Weizmann Institute, Rehovot, Israel)
- Fei Chen (Broad Institute, Boston, USA)
- Irit Sagi (Weizmann Institute, Rehovot, Israel)
- Charles Couturier (McGill, Montreal, Canada)
National:
- Robert Zeiser (Freiburg)
- Marco Prinz (Freiburg)
- Ian Frew (Freiburg)
- Jürgen Beck (Freiburg)
- Vidhja Ravi (Freiburg)
- Nicolas Neidert (Freiburg)
- Tobias Wertheimer (Freiburg)
- Daniel Kirschenbaum (DKFZ, Heidelberg)
* equal Contribution
- Tyler M, Gavish A, Barbolin C, Tschernichovsky R, Hoefflin R, Mints M, Puram S V, Tirosh I. The Curated Cancer Cell Atlas: comprehensive characterisation of tumours at single-cell resolution. Nature Cancer (2025)
- Greenwald AC*, Darnell NG*, Hoefflin R*, Simkin D, Gonzalez-Castro LN, Mount C, Hirsch D, Masashi N, Talpir T, Kedemi M, Goliand I, Medici G, Li B, Keren-Shaul H, Weller M, Addadi Y, Neidert MC, Suvà ML, Tirosh I. Integrative spatial analysis reveals a multi-layered organization of glioblastoma. Cell (2024)
Highlighted in Cancer Discovery “Reflections on Advances in Cancer Research in 2024”. - Harnik Y*, Yakubovsky O*, Hoefflin R*, Novoselsky R, Bahar Halpern K, Barkai T, Korem Kohanim Y, Egozi A, Golani O, Addadi Y, Kedmi M, Keidar Haran T, Levin Y, Savidor A, Keren-Shaul H, Pencovich N, Pery R, Shouval D, Tirosh I, Nachmany I, Itzkovitz S. (2024) A spatial expression atlas of the adult human proximal small intestine. Nature (2024)
- Schmidt D, Endres C, Hoefflin R, Andrieux G, Zwick M, Karantzelis N, Staehle HF, Vinnakota JM, Duquesne S, Mozaffari Jovein M, Pfeifer D, Becker H, Blazar BR, Zähringer A, Duyster J, Brummer T, Boerries M, Baumeister J, Shoumariyeh K, Li J, Green AR, Heidel FH, Tirosh I, Pahl HL, Leimkühler N, Köhler N, de Toledo MAS, Koschmieder S, Zeiser R. Oncogenic Calreticulin Induces Immune Escape by Stimulating TGFβ Expression and Regulatory T-cell Expansion in the Bone Marrow Microenvironment. Cancer Research (2024)
- Gavish A, Tyler M, Greenwald AC, Hoefflin R, Simkin D, Tschernichovsky R, Galili Darnell N, Somech E, Barbolin C, Antman T, Kovarsky D, Barrett T, Gonzalez Castro, LN, Halder D, Chanoch-Myers R, Laffy J, Mints M, Wider A, Tal R, Spitzer A, Hara T, Raitses-Gurevich M, Stossel C, Golan T, Tirosh A, Suvà ML, Puram SV, Tirosh I. (2023) Hallmarks of transcriptional intratumour heterogeneity across a thousand tumours. Nature (2023)
Highlighted in Nature, Cancer Cell, and Nature Reviews Genetics. - Hoefflin R, Harlander S, Abhari BA, Peighambari A, Adlesic M, Seidel P, Zodel K, Haug S, Göcmen B, Yong L, Lahrmann B, Grabe N, Heide D, Boerries M, Köttgen A, Heikenwalder M, Frew IJ. (2021) Therapeutic Effects of Inhibition of Sphingosine-1-Phosphate Signaling in HIF-2α Inhibitor-Resistant Clear Cell Renal Cell Carcinoma. Cancers (2021)
- Hoefflin R, Lazarou A, Hess ME, Reiser M, Wehrle J, Metzger P, Frey A, Becker H, Aumann K, Berner K, Boeker M, Buettner N, Dierks D, Duque-Afonso J, Eisenblaetter M, Erbes T, Fritsch R, Ge IX, Geißler AL, Grabbert M, Heeg S, Heiland DH, Hettmer S, Kayser G, Keller A, Kleiber A, Kutilina A, Mehmed L, Meiss F, Poxleitner P, Rawluk J, Ruf J, Schäfer H, Scherer F, Shoumariyeh S, Tzschach A, Peters C, Brummer T, Werner M, Duyster J, Lassmann S, Miething C, Boerries M, Illert AL, von Bubnoff N. (2021) Transitioning the Molecular Tumor Board from proof of concept to clinical routine: A German single-center analysis. Cancers (2021)
- Hoefflin R*, Harlander S*, Schäfer S, Metzger P, Kuo F, Schönenberger D, Adlesic M, Peighambari A, Seidel P, Chen CY, Consenza-Contreras M, Jud A, Lahrmann B, Grabe N, Heide D, Uhl FM, Chan TA, Duyster J, Zeiser R, Schell C, Heikenwalder M, Schilling O, Hakimi AA, Boerries M, Frew IJ. HIF-1α and HIF-2α differently regulate tumour development and inflammation of clear cell renal cell carcinoma in mice. Nature Communications (2020)
- Hoefflin R, Geißler AL, Fritsch R, Claus R, Wehrle J, Metzger P, Reiser M, Mehmed L, Fauth L, Heiland DH, Erbes T, Stock F, Csanadi A, Miething C, Weddeling B, Meiss F, von Bubnoff D, Dierks C, Ge I, Brass V, Heeg S, Schäfer H, Boeker M, Rawluk J, Botzenhart EM, Kayser G, Hettmer S, Busch H, Peters C, Werner M, Duyster J, Brummer T, Boerries M, Lassmann S, von Bubnoff N. Personalized Clinical Decision Making Through Implementation of a Molecular Tumor Board: A German Single-Center Experience. JCO Precision Oncology (2018)
- Hoefflin R, Lahrmann B, Warsow G, Hübschmann D, Spath C, Walter B, Chen X, Hofer L, Macher-Goeppinger S, Tolstov Y, Korzeniewski N, Duensing A, Grüllich C, Jäger D, Perner S, Schönberg G, Nyarangi-Dix J, Isaac S, Hatiboglu G, Teber D, Hadaschik B, Pahernik S, Roth W, Eils R, Schlesner M, Sültmann H, Hohenfellner M, Grabe N, Duensing S. Spatial niche formation but not malignant progression is a driving force for intratumoural heterogeneity. Nature Communications (2016)
| 2026 | Weizmann Institute Prize for Outstanding Achievements in Postdoctoral Research |
| 2025 | Fellowship of the BMBF funded Advanced Clinician-Scientist Program IMMediate |
| 2024 | Vincenz-Czerny award of the German Society of Hematology and Medical Oncology |
| 2022 - 2024 | Walter-Benjamin Program of the German Research Foundation |
| 2021 | Publication award of the Else-Kröner Fresenius Foundation |
| 2018 - 2021 | Else-Kröner Fresenius project funding: “Impact of HIF on the tumor microenvironment of clear cell renal cell carcinoma” |
| 2017 - 2021 | Fellowship of the Clinician-Scientist Program Berta-Ottenstein of the Medical Faculty, University Hospital Freiburg, Germany |
| 2017 | Medical doctorate award of the German Society of Hematology and Medical Oncology |
| 2014 | Fellowship in the US-program from the Medical Faculty of Heidelberg and Fellowship in the abroad program PROMOS from the German Academic Exchange Service |
| 2013 | Fellowship in the junior research program Talents in Medicine of the Heidelberg University, Germany |