INGELFINGER LABORATORY
Laboratory for Generative Single Cell ImmunologyAbout Us
The Laboratory for Generative Single Cell Immunology develops and applies experimental and computational single-cell technologies to transform diagnostics and therapeutics in cancer care.
Single-cell methodologies enable the interrogation of heterogeneous systems such as the human immune system and how they become dysregulated during disease. These technologies provide a powerful framework to identify novel biomarkers, reconstruct disease trajectories, and uncover mechanisms of immune escape. At the same time, high-dimensional single-cell data are inherently complex and often confounded by technical variability, posing fundamental challenges for interpretation, integration, and clinical translation.
Our lab addresses this challenge by combining next-generation single-cell and spatial immune profiling with generative machine-learning models. We develop and apply methods to resolve disease-relevant immune cell states, identify maladaptive immune trajectories underlying immune escape and therapy resistance, and quantify cell–cell interactions across space and time.
Working at the interface of systems immunology, computational biology, and translational oncology, we integrate experimental innovation and quantitative modeling to transform complex single-cell data into predictive and clinically actionable insight, supporting precision diagnostics and the rational design of next-generation immunotherapies
Research Topics
Our research is structured around two technology-driven focus areas, which we apply to address pressing translational questions in oncology and hematology.
1. Generative AI for Translational Research utilizing Antibody-based Single-Cell Technologies
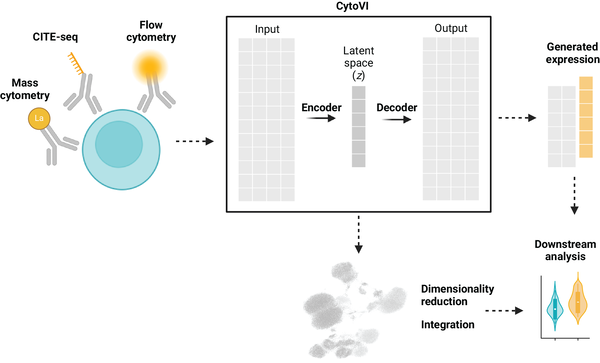
We develop generative machine-learning models for antibody-based single-cell technologies to enable robust, scalable, and clinically meaningful immune profiling (European Journal of Immunology, 2025). Antibody-based single cell assays such as high-dimensional flow cytometry or CITE-seq pose a unique opportunity to understand disease mechanisms across a large number of specimens (Nature, 2022), yet are limited by technical variability, low dimensionality, and poor comparability across panels, instruments, and time.
Our work focuses on deep probabilistic modeling to mitigate technical variability and computationally increase assay dimensionality through marker imputation and data harmonization (BioRxiv, 2025). Utilizing large archives of clinical cytometry data, we build foundation models of immune cell variability that capture disease-relevant immune states across large patient populations. These models are prospectively enriched with high-dimensional single-cell genomics measurements to generate insight about underlying transcriptional programs associated with disease and improve patient classification and longitudinal disease monitoring.
We are further extending these frameworks to incorporate additional emerging modalities, including subcellular protein colocalization and cellular morphology, enabling a novel perspective on cellular function. Thereby, we bridge research-grade technologies and clinical workflows in an interdisciplinary approach.
2. Spatiotemporal Dissection of Immune Dynamics in Cancer
We develop and apply space- and time-resolved single-cell genomics to dissect how immune cells infiltrate tumors, interact within distinct tissue niches, and become dysregulated within the tumor microenvironment (Cell, 2024). These approaches allow us to reconstruct immune cell state trajectories, and interaction networks across space and time, providing mechanistic insight into immune dysregulation.
By resolving how immune programs evolve within specific microenvironments, we aim to identify the cellular and molecular drivers of immune escape, therapy resistance, and treatment failure. This work establishes a dynamic, systems-level understanding of immune–tumor ecosystems and provides a mechanistic foundation for the rational design and optimization of immunotherapies (Cancer Cell, 2025).
Team

Principal Investigator
Head of Ingelfinger Lab
Florian Ingelfinger, PhD
florian.ingelfinger@uniklinik-freiburg.de
🆔 ORCID
✖ X

Open Positions

Our lab is always looking for highly motivated and curious talents who want to set the pace in a technology-focused, translational immunology lab.
We are a highly interdisciplinary team working at the interface of experimental immunology, single-cell biology, computational science, and oncology.
We welcome applications from interns, Master’s students, MD and MD/PhD students, PhD students, and postdoctoral researchers with backgrounds in immunology, molecular or cellular biology, bioinformatics, computational biology, computer science, or related disciplines. We offer wet lab, dry lab or hybrid projects. Please reach out with a short description of your interests and a CV.
The research group of Dr. Florian Ingelfinger, Centre for Translational Cell Research (ZTZ), at the Medical Center – University of Freiburg, Department of Hematology, Oncology and Stem Cell Transplantation, invites applications from
Master Students/Internships (m/f/d)
Starting date: as soon as possible
Project: Deep Generative Modeling of Immune Phenotypes in Acute Myeloid Leukemia
The research group of Dr. Florian Ingelfinger, Centre for Translational Cell Research (ZTZ) at the Medical Center – University of Freiburg, Department of Hematology, Oncology and Stem Cell Transplantation, invites applications from as soon as possible
PhD Candidates (m/f/d)
Project title: Spatio-temporal Mechanisms of Immune Escape in Brain Metastases
The position is funded within the Collaborative Research Centre CRC 1479 “OncoEscape – Mechanisms of Immune Escape in Cancer.”
Collaborators
International:
- Sarah Mundt (University Hospital Zurich, Zurich, Switzerland)
- Ido Amit (Weizmann Institute, Rehovot, Israel)
- Nir Yosef (Weizmann Institute, Rehovot, Israel)
- Bettina Schreiner (University Hospital Zurich, Zurich, Switzerland)
- Burkhard Becher (University of Zurich, Zurich, Switzerland)
Corinne Widmer (University Hospital Basel, Basel, Switzerland)
National:
- Tobias Wertheimer (Freiburg)
- Rouven Hoefflin (Freiburg)
- Daniel Kirschenbaum (DKFZ, Heidelberg)
- Robert Zeiser (Freiburg)
- Melanie Börries (Freiburg)
- Bertram Bengsch (Freiburg)
- Jesus Duque Afonso (Freiburg)
- Marco Prinz (Freiburg)
- Roman Sankowski (Freiburg)
- Melanie Sieder (Freiburg)
- Ian Frew (Freiburg)
- Heinz Wiendl (Freiburg)
- Ari Waisman (Johannes Gutenberg University, Mainz)
- Lisa Sevenich (University of Tübingen)
Publications
The full publication list can be found at google scholar. Below a selection:
Note: #, $ or * indicate that authors marked with the same symbols contributed equally to each study.
- Ingelfinger F*, Gerdes LA*, Kavaka V, Krishnarajah S, Friebel E, Galli E, Zwicky P, Furrer R, Peukert C, Dutertre CA, Eglseer KM, Ginhoux F, Flierl-Hecht A, Kümpfel T, De Feo D, Schreiner B, Mundt S, Kerschensteiner M, Hohlfeld R , Beltrán E#, Becher B#. (2022) Twin study reveals non-heritable immune perturbations in multiple sclerosis. Nature.
- Kirschenbaum D*, Xie K*, Ingelfinger F*, Katzenelenbogen Y*, Abadie K*, Look T, Sheban F, Phan TS, Li B, Zwicky P, Yofe I, David E, Mazuz K, Hou J, Chen Y, Shaim H, Shanley M, Becker S, Qian J, Colonna M, Ginhoux F, Rezvani K, Theis FJ, Yosef N, Weiss T, Weiner A#, Amit I#. (2024) Time-resolved single-cell transcriptomics defines immune trajectories in glioblastoma. Cell.
- Sheban F*, Truong SP*, Xie K*, Ingelfinger F*, Gur C, Shapir Y, Shapir I, Blecher-Gonen R, Yu C, Avellino R, Chalan P, Freitag K, Yofe I, Yutkin V, Boyeau P, Ergen C, Hong J, Mazuz K, Liu Y, Chen K, Dahan R, Kortylewski M, Yosef N, Weiner A, Amit I. (2025) ZEB2 is a master switch controlling the tumor-associated macrophage program. Cancer Cell.
- Andreadou M*, Ingelfinger F*, De Feo D$, Cramer T$, Tuzlak S$, Friebel E, Schreiner B, Eede P, Schneeberger S, Geesdorf M, Ridder F, Welsh C, Power L, Kirschenbaum D, Tyagarajan S, Greter M, Heppner F, Mundt S#, Becher B#. (2023) IL-12 sensing in neurons induces neuroprotective CNS tissue adaptation and attenuates neuroinflammation in mice. NatureNeuroscience.
- Ulutekin C*, Galli E*, Schreiner B, Khademi M, Callegari I, Piehl F, Sanderson N, Kirschenbaum D, Mundt S, Filippi M, Furlan R, Olsson T, Derfuss T, Ingelfinger F#, Becher B#. (2024) B cell depletion attenuates CD27 signaling of T helper cells in multiple sclerosis. Cell Reports Medicine.
- Ingelfinger F, Krishnarajah S, Kramer M, Utz SG, Galli E, Lutz M, Zwicky P, Akarca AU, Jurado NP, Ulutekin C, Bamert D, Widmer CC, Piccoli L, Sallusto F, Núñez NG, Marafioti T, Schneiter D, Opitz I, Lanzavecchia A, Jung HH, De Feo D, Mundt S, Schreiner B*, Becher B*.(2021) Single-cell profiling of myasthenia gravis identifies a pathogenic T cell signature. Acta Neuropathologica.
- Ingelfinger F, Kuiper KL, Ulutekin C, Rindlisbacher L, Mundt S, Gerdes LA, Smolders J, M van Luijn MM#, Becher B#. (2024) Twin study dissects CXCR3+ memory B cells as non-heritable feature in multiple sclerosis. Med.
- Ingelfinger F*, Sparano C*, Bamert D, Reyes-Leiva D, Sethi A, Rindlisbacher L, Zwicky P, Kreutmair S, Widmer C, Mundt S, Cortés-Vicente E, Tugues S, Becher B#, Schreiner B#. (2022) Azathioprine therapy induces selective NK cell Depletion and IFN-γ Deficiency predisposing to herpes virus reactivation. J Allergy Clin Immunol.
- Ingelfinger F*, Kramer M*, Lutz M, Widmer C, Piccoli L, Kreutmair S, Wertheimer T, Woodhall M, Waters P, Sallusto F, Lanzavecchia A, Mundt S, Becher B#, Schreiner B#. (2023)
Antibodies Produced by CLL Phenotype B Cells in Patients With Myasthenia Gravis Are Not Directed Against Neuromuscular Endplates. Neurol Neuroimmunol NeuroInflamm. - 1Rodov A*, Baniadam H*, Zeiser R, Amit I, Yosef N, Wertheimer T#, Ingelfinger F#. (2025) Towards the Next Generation of Data-Driven Therapeutics Using Spatially Resolved Single-Cell Technologies and Generative AI. Eur. J. Immunol.
Awards
2024 Marie Skłodowska-Curie Actions Postdoctoral Fellowship
2023 EMBO Postdoctoral fellowship
2023 Deutsche Gesellschaft für Muskelkranke Junior Researcher Award
2023 Pfizer Research Award
2022 Koshland Prize
2022 Neuromuscular Research Association Basel Grant
2022 Research Award Klinisches Neurozentrum
2020 MS Society Research Grant
2019 Studienstiftung des deutschen Volkes Fellowship